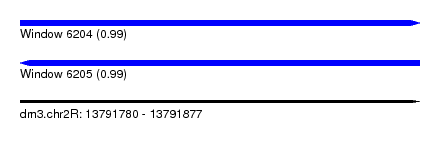
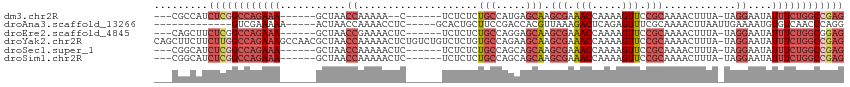
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,791,780 – 13,791,877 |
| Length | 97 |
| Max. P | 0.994383 |

| Location | 13,791,780 – 13,791,877 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.78 |
| Shannon entropy | 0.43633 |
| G+C content | 0.46137 |
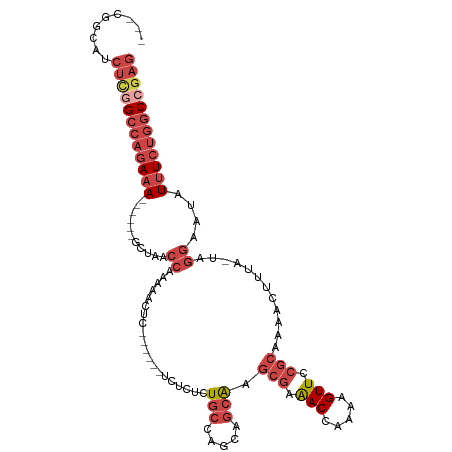
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -23.61 |
| Energy contribution | -25.15 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
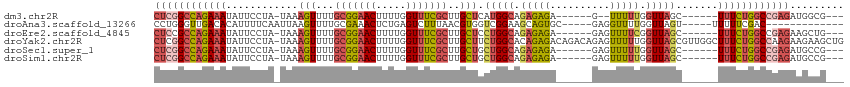
>dm3.chr2R 13791780 97 + 21146708 CUCGGCCAGAAAUAUUCCUA-UAAAGUUUUGCGGAACUUUUGGUUUCGCUUGCUCAUGGCAGAGAGA------G--UUUUUGGUUAGC------UUUCUGGCCGAGAUGGCG--- ((((((((((((..(..(((-.(((.(((((((((((.....)))))))((((.....)))).))))------.--))).)))..)..------))))))))))))......--- ( -38.90, z-score = -4.09, R) >droAna3.scaffold_13266 9484690 92 - 19884421 CCUGGGUUGACACAUUUUCAAUUAAGUUUUGCGAAACUCUGAGUCUUUAACGUGGUCGGAAGCAGUGC-----GAGGUUUUGGUUAGU-----UUUUUCGAC------------- .((..(((((.......)))))..)).....(((((..((((..((..(((.(.(((....)....))-----.).)))..)))))).-----..)))))..------------- ( -14.20, z-score = 2.18, R) >droEre2.scaffold_4845 8019360 99 + 22589142 CUCCGCCAGAAAUAUUCCUA-UAAAGUUUUGCGGAACUUUUGGUUUCGCUUGCUCCUGGCAGAGAGA------GAGUUUUCGGUUAGC------UUUCUGGCCGAGAAGCUG--- (((.(((((...(((....)-)).(((...(((((((.....)))))))..))).))))).)))...------.(((((((((((((.------...))))))))).)))).--- ( -34.90, z-score = -2.21, R) >droYak2.chr2R 5748802 114 - 21139217 CUCGGCCAGAAAUAUUCCUA-UAAAGUUUUGCGGAACUUUUGGUUUCGCUUGCUUCUGGCACAGAGACAGACAGAGUUUUUGGUUAGCGUUGGCUUUCUGGCCAAGAAGAAGCUG ((.(((((((((....((..-..((((...(((((((.....)))))))..))))(((((.(((((((.......))))))))))))....)).))))))))).))......... ( -37.90, z-score = -1.93, R) >droSec1.super_1 11286928 99 + 14215200 CUCGGCCAGAAAUAUUCCUA-UAAAGUUUUGCGGAACUUUUGGUUUCGCUUGCUGCUGGCAGAGAGA------GAGUUUUUGGUUAGC------UUUCUGGCCGAGAUGCCG--- ((((((((((((........-...(((...(((((((.....)))))))..)))((((((.(((((.------...))))).))))))------))))))))))))......--- ( -41.50, z-score = -4.28, R) >droSim1.chr2R 12534479 99 + 19596830 CUCGGCCAGAAAUAUUCCUA-UAAAGUUUUGCGGAACUUUUGGUUUCGCUUGCUGCUGGCAGAGAGA------GAGUUUUUGGUUAGC------UUUCUGGCCGAGAUGCCG--- ((((((((((((........-...(((...(((((((.....)))))))..)))((((((.(((((.------...))))).))))))------))))))))))))......--- ( -41.50, z-score = -4.28, R) >consensus CUCGGCCAGAAAUAUUCCUA_UAAAGUUUUGCGGAACUUUUGGUUUCGCUUGCUGCUGGCAGAGAGA______GAGUUUUUGGUUAGC______UUUCUGGCCGAGAUGCCG___ ((((((((((((............(((...(((((((.....)))))))..))).(((((.(((((..........))))).))))).......))))))))))))......... (-23.61 = -25.15 + 1.54)
| Location | 13,791,780 – 13,791,877 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.78 |
| Shannon entropy | 0.43633 |
| G+C content | 0.46137 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -17.14 |
| Energy contribution | -19.25 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
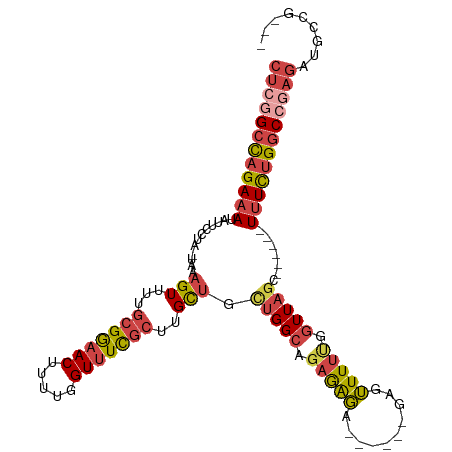
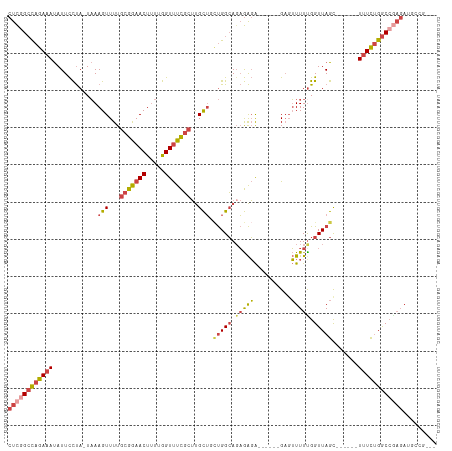
>dm3.chr2R 13791780 97 - 21146708 ---CGCCAUCUCGGCCAGAAA------GCUAACCAAAAA--C------UCUCUCUGCCAUGAGCAAGCGAAACCAAAAGUUCCGCAAAACUUUA-UAGGAAUAUUUCUGGCCGAG ---......((((((((((((------.....((.....--.------...(((......)))...(((.(((.....))).))).........-..))....)))))))))))) ( -30.20, z-score = -4.53, R) >droAna3.scaffold_13266 9484690 92 + 19884421 -------------GUCGAAAAA-----ACUAACCAAAACCUC-----GCACUGCUUCCGACCACGUUAAAGACUCAGAGUUUCGCAAAACUUAAUUGAAAAUGUGUCAACCCAGG -------------.........-----...............-----...(((.....(((.(((((...((....((((((....))))))..))...))))))))....))). ( -8.50, z-score = 1.36, R) >droEre2.scaffold_4845 8019360 99 - 22589142 ---CAGCUUCUCGGCCAGAAA------GCUAACCGAAAACUC------UCUCUCUGCCAGGAGCAAGCGAAACCAAAAGUUCCGCAAAACUUUA-UAGGAAUAUUUCUGGCGGAG ---.(((((.((.....))))------)))............------...((((((((((((...(((.(((.....))).)))....((...-.)).....)))))))))))) ( -27.40, z-score = -1.65, R) >droYak2.chr2R 5748802 114 + 21139217 CAGCUUCUUCUUGGCCAGAAAGCCAACGCUAACCAAAAACUCUGUCUGUCUCUGUGCCAGAAGCAAGCGAAACCAAAAGUUCCGCAAAACUUUA-UAGGAAUAUUUCUGGCCGAG .........((((((((((((.((..((((...((.......))..((..((((...))))..))))))......((((((......)))))).-..))....)))))))))))) ( -32.00, z-score = -2.67, R) >droSec1.super_1 11286928 99 - 14215200 ---CGGCAUCUCGGCCAGAAA------GCUAACCAAAAACUC------UCUCUCUGCCAGCAGCAAGCGAAACCAAAAGUUCCGCAAAACUUUA-UAGGAAUAUUUCUGGCCGAG ---......((((((((((((------.....((......((------.((..(((....)))..)).)).....((((((......)))))).-..))....)))))))))))) ( -30.20, z-score = -3.64, R) >droSim1.chr2R 12534479 99 - 19596830 ---CGGCAUCUCGGCCAGAAA------GCUAACCAAAAACUC------UCUCUCUGCCAGCAGCAAGCGAAACCAAAAGUUCCGCAAAACUUUA-UAGGAAUAUUUCUGGCCGAG ---......((((((((((((------.....((......((------.((..(((....)))..)).)).....((((((......)))))).-..))....)))))))))))) ( -30.20, z-score = -3.64, R) >consensus ___CGGCAUCUCGGCCAGAAA______GCUAACCAAAAACUC______UCUCUCUGCCAGCAGCAAGCGAAACCAAAAGUUCCGCAAAACUUUA_UAGGAAUAUUUCUGGCCGAG .........((((((((((((.................................(((.....))).(((.(((.....))).)))..................)))))))))))) (-17.14 = -19.25 + 2.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:53 2011