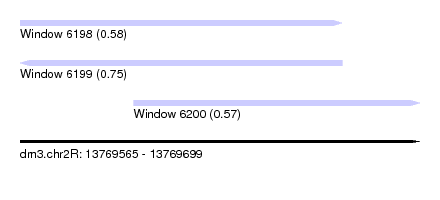
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,769,565 – 13,769,699 |
| Length | 134 |
| Max. P | 0.746958 |

| Location | 13,769,565 – 13,769,673 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.86 |
| Shannon entropy | 0.57372 |
| G+C content | 0.49297 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -13.95 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.578748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
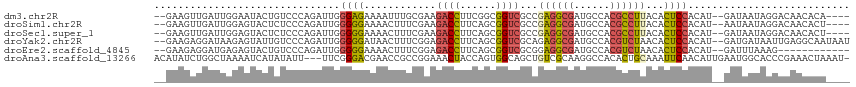
>dm3.chr2R 13769565 108 + 21146708 --GAAGUUGAUUGGAAUACUGUCCCAGAUUGGGAGAAAAUUUGCGAAGACCUUCGGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU--GAUAAUAGGACAACACA---- --...((((.(((.((...(.((((.....)))).)....)).)))...((((((((....)))))((((......)))).............--......))).))))...---- ( -28.30, z-score = -0.81, R) >droSim1.chr2R 12511467 108 + 19596830 --GAAGUUGAUUGGAGUACUCUCCCAGAUUGGGGGAAAACUUUCGAAGACCUUCAGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU--AAUAAUAGGACAACACU---- --...((((..((((((..((((((.....))))))......(((..((((......))))..)))((((......))))....))))))(..--.......)..))))...---- ( -32.70, z-score = -2.15, R) >droSec1.super_1 11265154 108 + 14215200 --GAAGUUGAUUGGAGUACUCUCCCAGAUUGGGGGAAAACUUUCGAAGACCUUCAGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU--GAUAAUAGGACAACACU---- --...((((..((((((..((((((.....))))))......(((..((((......))))..)))((((......))))....)))))).((--....))....))))...---- ( -33.20, z-score = -1.92, R) >droYak2.chr2R 5725903 112 - 21139217 --GAAGAGGAUAAGAGUAUUGUCCCAGAUUGGGGGAUAACUUUCGGAGACCUUCAGCGGUCGCAGAGGCGAUGCCACGUCUAACACUCCACAU--GAUGAUAAUUGAGGCAAUAAU --((((.(..(..(((..(((((((.......)))))))..)))..)..)))))....(((((....)))))(((.((..((.((.((.....--)))).))..)).)))...... ( -28.20, z-score = -0.25, R) >droEre2.scaffold_4845 7997187 100 + 22589142 --GAAGAGGAUGAGAGUACUGUCCCAGAUUGGGGGAAAACUUUCGGAGACCUUCAGCGGUCGCGGAGGCGAUGCCACGUCUAACACUCCACAU--GAUUUAAAG------------ --.........((((((..(.((((.....)))).)..))))))((((.(((((.((....))))))).((((...)))).....))))....--.........------------ ( -27.70, z-score = -0.19, R) >droAna3.scaffold_13266 9463947 112 - 19884421 ACAUAUCUGGCUAAAAUCAUAUAUU---UUCGGGACGAACCGCCGGAAACUACCAGUGGCAGCUGUCGCAAGGCCACACUGCAAAUUCAACAUUGAAUGGCACCCGAAACUAAAU- ........................(---((((((.......((((....).....(((((....)))))..))).....(((..(((((....))))).))))))))))......- ( -27.10, z-score = -1.39, R) >consensus __GAAGUUGAUUAGAGUACUGUCCCAGAUUGGGGGAAAACUUUCGAAGACCUUCAGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU__GAUAAUAGGACAACACA____ ...............................((((............((((......))))...((((((......))))))...))))........................... (-13.95 = -14.15 + 0.20)
| Location | 13,769,565 – 13,769,673 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.86 |
| Shannon entropy | 0.57372 |
| G+C content | 0.49297 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -18.07 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13769565 108 - 21146708 ----UGUGUUGUCCUAUUAUC--AUGUGGAGUGUAAGGCGUGGCAUCGCCUCGGCGACCGCCGAAGGUCUUCGCAAAUUUUCUCCCAAUCUGGGACAGUAUUCCAAUCAACUUC-- ----.(((((((((((.....--.((.((((......(((.((....((((((((....)))).)))))).))).......))))))...))))))))))).............-- ( -31.92, z-score = -1.49, R) >droSim1.chr2R 12511467 108 - 19596830 ----AGUGUUGUCCUAUUAUU--AUGUGGAGUGUAAGGCGUGGCAUCGCCUCGGCGACCGCUGAAGGUCUUCGAAAGUUUUCCCCCAAUCUGGGAGAGUACUCCAAUCAACUUC-- ----...((((..........--...(((((((...((((......))))((((.((((......)))).))))...(((((((.......))))))))))))))..))))...-- ( -33.56, z-score = -1.63, R) >droSec1.super_1 11265154 108 - 14215200 ----AGUGUUGUCCUAUUAUC--AUGUGGAGUGUAAGGCGUGGCAUCGCCUCGGCGACCGCUGAAGGUCUUCGAAAGUUUUCCCCCAAUCUGGGAGAGUACUCCAAUCAACUUC-- ----...((((..........--...(((((((...((((......))))((((.((((......)))).))))...(((((((.......))))))))))))))..))))...-- ( -33.56, z-score = -1.46, R) >droYak2.chr2R 5725903 112 + 21139217 AUUAUUGCCUCAAUUAUCAUC--AUGUGGAGUGUUAGACGUGGCAUCGCCUCUGCGACCGCUGAAGGUCUCCGAAAGUUAUCCCCCAAUCUGGGACAAUACUCUUAUCCUCUUC-- .....................--....((((((((....((((..((((....))))))))....((....(....)....))(((.....)))..))))))))..........-- ( -22.70, z-score = 0.52, R) >droEre2.scaffold_4845 7997187 100 - 22589142 ------------CUUUAAAUC--AUGUGGAGUGUUAGACGUGGCAUCGCCUCCGCGACCGCUGAAGGUCUCCGAAAGUUUUCCCCCAAUCUGGGACAGUACUCUCAUCCUCUUC-- ------------.........--..(((((((((((....)))))....))))))((((......))))...((.(((.....(((.....))).....))).)).........-- ( -23.50, z-score = 0.22, R) >droAna3.scaffold_13266 9463947 112 + 19884421 -AUUUAGUUUCGGGUGCCAUUCAAUGUUGAAUUUGCAGUGUGGCCUUGCGACAGCUGCCACUGGUAGUUUCCGGCGGUUCGUCCCGAA---AAUAUAUGAUUUUAGCCAGAUAUGU -......(((((((.(((((....(((.......)))..)))))...((((..((((((...((......))))))))))))))))))---)((((.((........)).)))).. ( -30.20, z-score = -0.45, R) >consensus ____UGUGUUGUCCUAUUAUC__AUGUGGAGUGUAAGGCGUGGCAUCGCCUCGGCGACCGCUGAAGGUCUCCGAAAGUUUUCCCCCAAUCUGGGACAGUACUCCAAUCAACUUC__ ..........................(((((((.(((((((((..((((....)))))))).....)))))............(((.....)))....)))))))........... (-18.07 = -17.72 + -0.36)

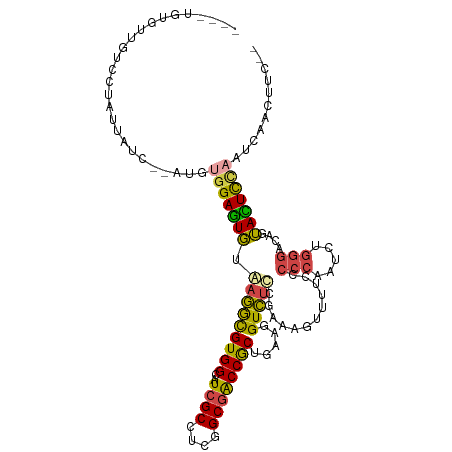

| Location | 13,769,603 – 13,769,699 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 64.91 |
| Shannon entropy | 0.61218 |
| G+C content | 0.45428 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -9.22 |
| Energy contribution | -9.67 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13769603 96 + 21146708 UUGCGAAGACCUUCGGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU--GAU----AAUAGGACAACACA----------AGAUUUACA---GAAAUUAACACGUAAUU- (((((....((((((((....)))))((((......)))).............--...----...)))........----------.(((((...---.)))))....)))))..- ( -20.40, z-score = -1.32, R) >droSim1.chr2R 12511505 96 + 19596830 UUUCGAAGACCUUCAGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU--AAU----AAUAGGACAACACU----------GGAUUUACA---GGAAAACACACGUAAUU- ..(((..((((......))))..)))((((......))))((((..(((....--...----....))).....((----------(......))---)..........))))..- ( -20.40, z-score = -1.54, R) >droSec1.super_1 11265192 96 + 14215200 UUUCGAAGACCUUCAGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU--GAU----AAUAGGACAACACU----------AGAUUUACA---GGAAUACACACGUAAUU- ..(((..((((......))))..)))((((......))))......(((...(--((.----..(((.......))----------)...)))..---)))..............- ( -18.40, z-score = -1.08, R) >droYak2.chr2R 5725941 113 - 21139217 UUUCGGAGACCUUCAGCGGUCGCAGAGGCGAUGCCACGUCUAACACUCCACAU--GAUGAUAAUUGAGGCAAUAAUUUAUUUACGGAAAUUAACGAUGGAAAAACGCUGAGAAUC- .((((....)..(((((((((((....))))).((((((.(((...(((....--(((((..((((...))))...)))))...)))..)))))).))).....)))))))))..- ( -26.30, z-score = -1.06, R) >droEre2.scaffold_4845 7997225 91 + 22589142 UUUCGGAGACCUUCAGCGGUCGCGGAGGCGAUGCCACGUCUAACACUCCACAU--GAU-----UUAAAG------------------GAUUAACAAUGGAAAAACGCUUAGAAUCU .(((...((((......))))(((.(((((......))))).....((((..(--(((-----(.....------------------)))))....))))....)))...)))... ( -19.50, z-score = -0.18, R) >droAna3.scaffold_13266 9463984 99 - 19884421 CGCCGGAAACUACCAGUGGCAGCUGUCGCAAGGCCACACUGCAAAUUCAACAUUGAAUGGCACCCGAAACUAAAUA-----------AAUUUAUUCU-UAAAACUUCACAU----- .((((....).....(((((.((....))...))))).......(((((....))))))))...............-----------..........-.............----- ( -18.90, z-score = -0.99, R) >consensus UUUCGAAGACCUUCAGCGGUCGCCGAGGCGAUGCCACGCCUUACACUCCACAU__GAU____AAUAAGACAACACU__________AGAUUUACA___GAAAAACACACAUAAUU_ .......((((......))))...((((((......)))))).......................................................................... ( -9.22 = -9.67 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:49 2011