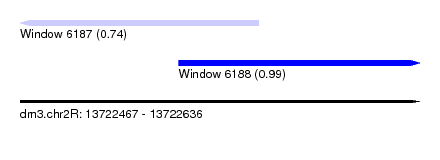
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,722,467 – 13,722,636 |
| Length | 169 |
| Max. P | 0.986483 |

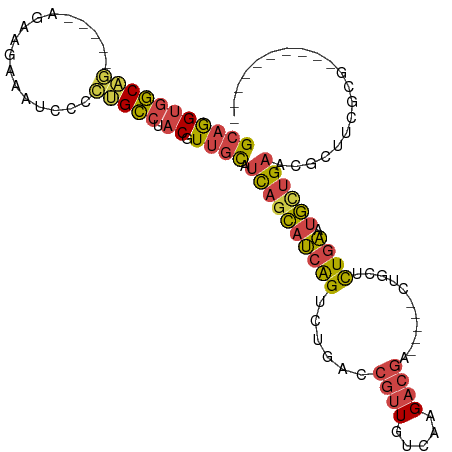
| Location | 13,722,467 – 13,722,568 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.55627 |
| G+C content | 0.52469 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
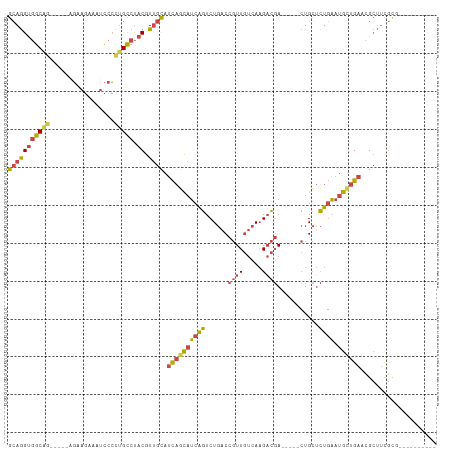
>dm3.chr2R 13722467 101 - 21146708 GCAGGUGGCAG-----AGAAGAAAUCCCCUGCCUACGUUGCAUCAGCAUCAGUCUGACCGUUGUCAAGACGA-----CUGCUCUGAAUGCUGAUCGCUUUGUGAGGGGAGA-- ((.....))..-----........((((((...((((..(((((((((((((...(.(.(((((....))))-----).)).))).)))))))).))..))))))))))..-- ( -35.60, z-score = -1.24, R) >droWil1.scaffold_180745 2164718 98 - 2843958 GGAAGCAACGACAACGACAACAACAUCGUCGUCCACAUCGUCGUCGUCGUGGUCU-UCUCUUCCUACGUUCA----UAUGUAUUUUAUAAUGAAAGAACCUUG---------- (((((....(((.(((((.........))))).(((..((....))..)))))).-...)))))....((((----((((.....))).))))).........---------- ( -19.80, z-score = -0.48, R) >droYak2.chr2R 5678480 96 + 21139217 GCAGGUGGC-------AGUAGAAAUCCCCUGCCUACGUUGCAUCAGCAUCAGUCUGACCGUUGUCAAGACGACUGCUCUGCUCUGAAUGCUGAUCGCUUUGGU---------- (((((((((-------((..(....)..))))).)).))))(((((((((((...(((.(((((....))))).).))....))).)))))))).........---------- ( -32.80, z-score = -1.57, R) >droEre2.scaffold_4845 7949769 97 - 22589142 GCAGGUGGCAGAAGGCAGUAGAAAUCCCCUGCCUACGUUGCAUCAGCAUCAGUCUGACCGUUGUCAAGACGA-----CUGCUCUGAAUGCUGAUCGCUUCGC----------- (((((((((((.((((((..(....)..))))))...))))(((((((((((...(.(.(((((....))))-----).)).))).))))))))))))).))----------- ( -37.00, z-score = -2.47, R) >droSec1.super_1 11218669 103 - 14215200 GCAGGUGGCAG-----AGAAGAAAUCCCCUGCCUACGUUGCAUCAGCAUCAGUCUGACCGUUGUCAAGACGA-----CUGCUCUGAAUGUUGAACGCUUCGCGAGAGGGGAUG (((((((((((-----.(.(....).).))))).)).)))).((((((((((...(.(.(((((....))))-----).)).))).))))))).(.((((....)))).)... ( -35.90, z-score = -1.41, R) >droSim1.chr2R 12462631 103 - 19596830 GCAGGUGGCAG-----CGAAGAAAUCCCCUGCCUACGUUGCAUCAGCAUCAGUCUGACCGUUGUCAAGACGA-----CUGCUCUGAAUGUUGAACGCUUCGCGAGAGGGGAUG (((((((((((-----.(.(....).).))))).)).)))).((((((((((...(.(.(((((....))))-----).)).))).))))))).(.((((....)))).)... ( -35.70, z-score = -1.14, R) >consensus GCAGGUGGCAG_____AGAAGAAAUCCCCUGCCUACGUUGCAUCAGCAUCAGUCUGACCGUUGUCAAGACGA_____CUGCUCUGAAUGCUGAACGCUUCGCG__________ (((((((((((.................))))).)).)))).((((((((((........((((....))))..........)))).)))))).................... (-15.30 = -15.50 + 0.20)

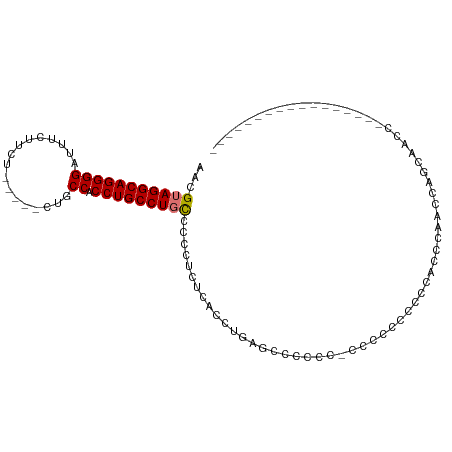
| Location | 13,722,534 – 13,722,636 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.39 |
| Shannon entropy | 0.51771 |
| G+C content | 0.64571 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -16.84 |
| Energy contribution | -16.91 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13722534 102 + 21146708 AACGUAGGCAGGGGAUUUCUUCU-----CUGCCACCUGCCUGCCCCCUCUCACCUGAGCUACACCCCCCUCCCCCCCCCACCGCGCUGCCCCUGCGCACCAGCAAUC ...(((((((((((.........-----...)).)))))))))...(((......)))........................((((.......)))).......... ( -27.40, z-score = -2.38, R) >droEre2.scaffold_4845 7949827 101 + 22589142 AACGUAGGCAGGGGAUUUCUACUGCCUUCUGCCACCUGCCUCUGCCCGCUCACCUGGGCCCCCUGCCCCCUGCCCCCUUUCCCACCUUCCCACUGGCAAUC------ ...(..((((((((.........((.....))...........(((((......)))))))))))))..)((((....................))))...------ ( -28.25, z-score = -1.18, R) >droSec1.super_1 11218738 82 + 14215200 AACGUAGGCAGGGGAUUUCUUCU-----CUGCCACCUGCCUGCCCCCUCUCACCCGUGCCCCCC---CUCCCACUAACCAACCAGCAAUC----------------- ...(((((((((((.........-----...)).))))))))).....................---.......................----------------- ( -18.30, z-score = -1.56, R) >droSim1.chr2R 12462700 84 + 19596830 AACGUAGGCAGGGGAUUUCUUCG-----CUGCCACCUGCCUGCCCC-UCUCACCUGAGCCCCCCUCCCUCCCUCUACCCAACCAGCAAUC----------------- ...(((((((((((.........-----...)).)))))))))..(-((......)))................................----------------- ( -20.10, z-score = -1.49, R) >consensus AACGUAGGCAGGGGAUUUCUUCU_____CUGCCACCUGCCUGCCCCCUCUCACCUGAGCCCCCC_CCCCCCCCCCACCCAACCAGCAACC_________________ ...(((((((((((.................)).)))))))))................................................................ (-16.84 = -16.91 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:40 2011