
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,683,409 – 13,683,520 |
| Length | 111 |
| Max. P | 0.619661 |

| Location | 13,683,409 – 13,683,520 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 126 |
| Reading direction | forward |
| Mean pairwise identity | 68.99 |
| Shannon entropy | 0.58781 |
| G+C content | 0.42712 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -15.25 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
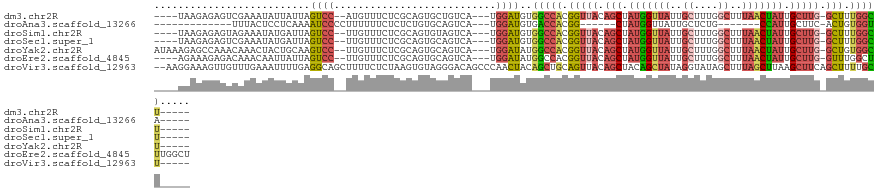
>dm3.chr2R 13683409 111 + 21146708 ----UAAGAGAGUCGAAAUAUUAUUAGUCC--AUGUUUCUCGCAGUGCUGUCA---UGGAUGUGGCCACGGUUACAGCUAUGGUUAUUGCUUUGGCUUUAACUAUUGCUUG-GCUUUGGCU----- ----......................((((--(((......(((....)))))---)))))..(((((.(((((.(((.(((((((..((....))..))))))).)))))-))).)))))----- ( -32.80, z-score = -1.39, R) >droAna3.scaffold_13266 9381186 91 - 19884421 -------------UUUACUCCUCAAAAUCCCCUUUUUUCUCUCUGUGCAGUCA---UGGAUGUGACCACGG------CUAUGGUUAUUGCUCUG-------CCAUUGCUUC-ACUGUGGUA----- -------------............................((((((....))---))))....(((((((------(.(((((.........)-------)))).)))..-...))))).----- ( -16.50, z-score = 0.03, R) >droSim1.chr2R 12423386 111 + 19596830 ----UAAGAGAGUAGAAAUAUGAUUAGUCC--UUGUUUCUCGCAGUGUAGUCA---UGGAUGUGGCCACGGUUACAGCUAUGGUUAUUGCUUUGGCUUUAACUAUUGCUUG-GCUUUGGCU----- ----.........(((((((.((....)).--.)))))))(((((((....))---)...))))((((.(((((.(((.(((((((..((....))..))))))).)))))-))).)))).----- ( -30.40, z-score = -0.65, R) >droSec1.super_1 11180343 111 + 14215200 ----UAAGAGAGUCGAAAUAUGAUUAGUCC--UUGUUUCUCGCAGUGCAGUCA---UGGAUGUGGCCACGGUUACAGCUAUGGUUAUUGCUUUGGCUUUAACUAUUGCUUG-GCUUUGGCU----- ----....((((((((((((.((....)).--.))))))..((((((....((---(((.((((((....)))))).)))))((((..((....))..))))))))))..)-)))))....----- ( -31.90, z-score = -0.85, R) >droYak2.chr2R 5638991 115 - 21139217 AUAAAGAGCCAAACAAACUACUGCAAGUCC--UUGUUUCUCGCAGUGCAGUCA---UGGAUAUGGCCACGGUUACAGCUAUGGUUAUUGCUUUGGCUUUAACUAUUGCUUG-GCUGUGGCU----- ........(((.......((((((.((...--......)).))))))......---)))....(((((((((((.(((.(((((((..((....))..))))))).)))))-)))))))))----- ( -38.82, z-score = -2.26, R) >droEre2.scaffold_4845 7910985 116 + 22589142 ----AGAAAGAGACAAACAAUUAUUAGUCC--UUGUUUCUCGCAGUGCAGUCA---UGGAUAUGGCCACGGUUACAGCUAUGGUUAUUGCUUUGGCUUUAACUAUUGCUUG-GUUUGGCUUUGGCU ----.(..((((((((((........))..--))))))))..).((.((....---)).))..(((((.(((((((((.(((((((..((....))..))))))).)).))-...))))).))))) ( -30.10, z-score = -0.33, R) >droVir3.scaffold_12963 19183327 119 + 20206255 --AAGGAAAGUUGUUUGAAAUUUUGAGGCAGCUUUUCUCUAAGUGUAGGGACAGCCCAACUACAGCUGCAGUUACAGCUACAGCUAUAGGUAUAGCUUUAGCUUAAGCUUCAGCUUUUGCU----- --.(((((((((((((.(.....).)))))))))))))....((((.(((....))).)).))(((((.((((..(((((.((((((....)))))).)))))..)))).)))))......----- ( -40.80, z-score = -1.92, R) >consensus ____UAAGAGAGUCGAAAUAUUAUUAGUCC__UUGUUUCUCGCAGUGCAGUCA___UGGAUGUGGCCACGGUUACAGCUAUGGUUAUUGCUUUGGCUUUAACUAUUGCUUG_GCUUUGGCU_____ ..........................((((........((........)).......))))..(((((.(((...(((.(((((((..((....))..))))))).)))...))).)))))..... (-15.25 = -16.13 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:37 2011