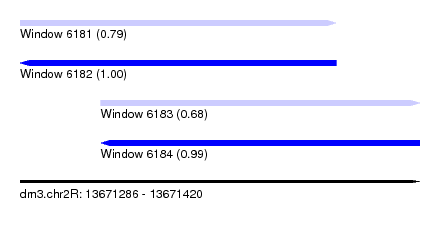
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,671,286 – 13,671,420 |
| Length | 134 |
| Max. P | 0.999472 |

| Location | 13,671,286 – 13,671,392 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Shannon entropy | 0.31188 |
| G+C content | 0.42585 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -19.25 |
| Energy contribution | -18.93 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13671286 106 + 21146708 ------------UCUCGCUGACUGUUCCAUUGAACAA-CUGACUAGAUGCAGCAUAGCGCUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAG ------------((.((.((..(((((....))))).-.((.(.((((((.(((((((...(((.((((....)))).)).)...))))))).).))))).).))....-..)).)))). ( -21.60, z-score = -0.65, R) >droEre2.scaffold_4845 7899183 106 + 22589142 ------------UCUCGCUGACUGUUCCAUUGAACAA-CUGACUAGAUGCAGCAUAGCGCUCUUUAAAAGCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAG ------------((.((.((..(((((....))))).-.((.(.((((((.((((((((((.......)))((........))..))))))).).))))).).))....-..)).)))). ( -24.20, z-score = -1.11, R) >droYak2.chr2R 5627091 106 - 21139217 ------------UCUCGCUGAGUGUUCCAUUGAACAA-CUGACUAGAUGCAGCAUAGCGAUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACAGAG ------------.....(((..(((((....))))).-.((.(.((((((.((((((((((.((.((((....)))).)).))).))))))).).))))).).))....-.....))).. ( -24.40, z-score = -1.47, R) >droSec1.super_1 11168759 106 + 14215200 ------------UCUCGCUGACUGUUCCAUUGAACAA-CUGACUAGAUGCAGCAUAGCGCUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAG ------------((.((.((..(((((....))))).-.((.(.((((((.(((((((...(((.((((....)))).)).)...))))))).).))))).).))....-..)).)))). ( -21.60, z-score = -0.65, R) >droSim1.chr2R 12409843 106 + 19596830 ------------UCUCGCUGACUGUUCCAUUGAACAA-CUGACUAGAUGCAGCAUAGCGCUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAG ------------((.((.((..(((((....))))).-.((.(.((((((.(((((((...(((.((((....)))).)).)...))))))).).))))).).))....-..)).)))). ( -21.60, z-score = -0.65, R) >droAna3.scaffold_13266 9368706 106 - 19884421 ------------UCUUGUUGACUGUUCCAUUGAACAA-CUGACUAGAUGCAGCAUAGCGCUCUCAAAAUUCGUUUUCCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-AACAACAGAU ------------..((((((..(((((....))))).-.((.(.((((((.(((((((............((((....))))...))))))).).))))).).))....-..)))))).. ( -25.66, z-score = -2.49, R) >dp4.chr3 16483205 105 + 19779522 -------------UCUGUUGACUGUUCCAUUGAAUAA-CUGACUAGAUGCAGCAUAGCGUUCUCAAAGAUCGCUUUUCAACUUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACAGAU -------------(((((((..(((((....))))).-.((.(.((((((.(((((((......((((....)))).........))))))).).))))).).))....-..))))))). ( -26.86, z-score = -2.88, R) >droPer1.super_4 6883662 105 - 7162766 -------------UCUGUUGACUGUUCCAUUGAAUAA-CUGACUAGAUGCAGCAUAGCGUUCUCAAAGAUCGCUUUUCAACUUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACAGAU -------------(((((((..(((((....))))).-.((.(.((((((.(((((((......((((....)))).........))))))).).))))).).))....-..))))))). ( -26.86, z-score = -2.88, R) >droWil1.scaffold_180700 4557866 101 + 6630534 ------------UAUUGUCGAUUGCUCUAUUGAAUAAACUGACUAGAUGCAGCAUAGCGCUUUUCUAAAAAUAUU------AUCAGCUAUGCCGACAUCUUGCCAAUUCGAAAAACAGA- ------------...(((((..(((((((.(.(......).).)))).)))(((((((.................------....))))))))))))......................- ( -19.40, z-score = -0.88, R) >droVir3.scaffold_12875 16937651 106 + 20611582 ------------UUUUGUUGAUUGUUCCAUUGAAUAA-CUGACUAGAUGCAGCAUAGCGCUAUAAAAAUGCGCUUUCAAACGUCAGCUAUGCCGACAUCUUGCCAAUUG-AACAACAGAU ------------.(((((((.((((((....))))))-.((.(.((((((.(((((((((.........))((........))..))))))).).))))).).))....-..))))))). ( -26.40, z-score = -2.09, R) >droMoj3.scaffold_6496 15801352 102 - 26866924 ---------------UGUUGACUGUUCCAUUGAAUAA-CUGACUAGAUGCAGCAUAGCGCUAU-GCAAUGCGCUUUGAAACGUCAGCUAUGCCGACAUCUUGCCAAUUC-AACAAUAGAU ---------------((((((.(((((....))))).-.((.(.((((((.(((((((((...-.....))....(((....)))))))))).).))))).).))..))-))))...... ( -27.00, z-score = -1.72, R) >droGri2.scaffold_15245 16476647 117 + 18325388 UUUCAGGUAUGGAUAUGUUGAGUGUUCCAUUGAAUAA-CUGACUAGAUGCAGCAUAGCGCUAU-AAGAUGCGCUUUCAAACGUCAGCUAUGCCGACAUCUUGCCAAUUC-ACCAACAGAU .....(((((((((((.....))).))))).((((..-.((.(.((((((.(((((((((...-.....))((........))..))))))).).))))).).))))))-)))....... ( -27.30, z-score = -0.30, R) >consensus ____________UCUUGUUGACUGUUCCAUUGAACAA_CUGACUAGAUGCAGCAUAGCGCUCUUAAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU_ACCAACAGAU ..............(((.((..(((((....)))))...((.(.((((((.(((((((...........................))))))).).))))).).)).......)).))).. (-19.25 = -18.93 + -0.32)



| Location | 13,671,286 – 13,671,392 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.87 |
| Shannon entropy | 0.31188 |
| G+C content | 0.42585 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -32.01 |
| Energy contribution | -31.98 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.31 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13671286 106 - 21146708 CUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUCAG-UUGUUCAAUGGAACAGUCAGCGAGA------------ .((((((((.-.(((((((.(((((.(((((((((...((((....)))).....(.....))))))))))))))).)))))-))..)))))))).............------------ ( -41.80, z-score = -4.11, R) >droEre2.scaffold_4845 7899183 106 - 22589142 CUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGCUUUUAAAGAGCGCUAUGCUGCAUCUAGUCAG-UUGUUCAAUGGAACAGUCAGCGAGA------------ .((((((((.-.(((((((.(((((.(((((((((....(((....)))((((.....)))))))))))))))))).)))))-))..)))))))).............------------ ( -46.20, z-score = -5.45, R) >droYak2.chr2R 5627091 106 + 21139217 CUCUGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAUCGCUAUGCUGCAUCUAGUCAG-UUGUUCAAUGGAACACUCAGCGAGA------------ (((.((((((-.(((((((.(((((.(((((((((....(((....)))((((.....)))))))))))))))))).)))))-))((((....)))))).))))))).------------ ( -43.70, z-score = -4.90, R) >droSec1.super_1 11168759 106 - 14215200 CUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUCAG-UUGUUCAAUGGAACAGUCAGCGAGA------------ .((((((((.-.(((((((.(((((.(((((((((...((((....)))).....(.....))))))))))))))).)))))-))..)))))))).............------------ ( -41.80, z-score = -4.11, R) >droSim1.chr2R 12409843 106 - 19596830 CUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUCAG-UUGUUCAAUGGAACAGUCAGCGAGA------------ .((((((((.-.(((((((.(((((.(((((((((...((((....)))).....(.....))))))))))))))).)))))-))..)))))))).............------------ ( -41.80, z-score = -4.11, R) >droAna3.scaffold_13266 9368706 106 + 19884421 AUCUGUUGUU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGGAAAACGAAUUUUGAGAGCGCUAUGCUGCAUCUAGUCAG-UUGUUCAAUGGAACAGUCAACAAGA------------ .(((((((..-.(((((((.(((((.(((((((((...((((....)))).....(....).)))))))))))))).)))))-))...))))))).............------------ ( -39.60, z-score = -4.34, R) >dp4.chr3 16483205 105 - 19779522 AUCUGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGAAGUUGAAAAGCGAUCUUUGAGAACGCUAUGCUGCAUCUAGUCAG-UUAUUCAAUGGAACAGUCAACAGA------------- .((((((((.-.(((((((.(((((.(((((((((....(((....)))(.((.....)).))))))))))))))).)))))-)).(((....)))...))))))))------------- ( -39.40, z-score = -4.59, R) >droPer1.super_4 6883662 105 + 7162766 AUCUGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGAAGUUGAAAAGCGAUCUUUGAGAACGCUAUGCUGCAUCUAGUCAG-UUAUUCAAUGGAACAGUCAACAGA------------- .((((((((.-.(((((((.(((((.(((((((((....(((....)))(.((.....)).))))))))))))))).)))))-)).(((....)))...))))))))------------- ( -39.40, z-score = -4.59, R) >droWil1.scaffold_180700 4557866 101 - 6630534 -UCUGUUUUUCGAAUUGGCAAGAUGUCGGCAUAGCUGAU------AAUAUUUUUAGAAAAGCGCUAUGCUGCAUCUAGUCAGUUUAUUCAAUAGAGCAAUCGACAAUA------------ -((((((....((((((((.(((((.(((((((((....------.................)))))))))))))).))))))))....)))))).............------------ ( -31.90, z-score = -3.65, R) >droVir3.scaffold_12875 16937651 106 - 20611582 AUCUGUUGUU-CAAUUGGCAAGAUGUCGGCAUAGCUGACGUUUGAAAGCGCAUUUUUAUAGCGCUAUGCUGCAUCUAGUCAG-UUAUUCAAUGGAACAAUCAACAAAA------------ .(((((((..-.(((((((.(((((.(((((((((....(((....)))((.........)))))))))))))))).)))))-))...))))))).............------------ ( -36.50, z-score = -4.16, R) >droMoj3.scaffold_6496 15801352 102 + 26866924 AUCUAUUGUU-GAAUUGGCAAGAUGUCGGCAUAGCUGACGUUUCAAAGCGCAUUGC-AUAGCGCUAUGCUGCAUCUAGUCAG-UUAUUCAAUGGAACAGUCAACA--------------- .(((((((..-.(((((((.(((((.(((((((((((.((((....))))))..((-...)))))))))))))))).)))))-))...)))))))..........--------------- ( -39.80, z-score = -4.53, R) >droGri2.scaffold_15245 16476647 117 - 18325388 AUCUGUUGGU-GAAUUGGCAAGAUGUCGGCAUAGCUGACGUUUGAAAGCGCAUCUU-AUAGCGCUAUGCUGCAUCUAGUCAG-UUAUUCAAUGGAACACUCAACAUAUCCAUACCUGAAA .((((((((.-.(((((((.(((((.(((((((((....(((....)))((.....-...)))))))))))))))).)))))-))..))))))))......................... ( -38.40, z-score = -3.13, R) >consensus AUCUGUUGGU_AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUCAG_UUAUUCAAUGGAACAGUCAACAAGA____________ .(((((((......(((((.(((((.(((((((((....(((....)))(...........))))))))))))))).)))))......)))))))......................... (-32.01 = -31.98 + -0.03)

| Location | 13,671,313 – 13,671,420 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Shannon entropy | 0.56165 |
| G+C content | 0.45961 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -15.13 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13671313 107 + 21146708 GACUAGAUGCAGCAUAGCGCUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAGUU-----GAUAUACGAUCGAUCCGACAUGGACG (.(.((((((.(((((((...(((.((((....)))).)).)...))))))).).))))).).).....-.(((.((((.((-----(((.....)))))))))...)))... ( -25.40, z-score = -1.28, R) >droEre2.scaffold_4845 7899210 107 + 22589142 GACUAGAUGCAGCAUAGCGCUCUUUAAAAGCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAGGU-----GCUAUACGAUCGAUCCGACAUGGCUC ....((((((.((((((((((.......)))((........))..))))))).).))))).((((...(-(((......)))-----).....((.......))...)))).. ( -25.80, z-score = -0.39, R) >droYak2.chr2R 5627118 107 - 21139217 GACUAGAUGCAGCAUAGCGAUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACAGAGGU-----GCUAUACGAUCGAGCCGACAUGGCUC (.(.((((((.((((((((((.((.((((....)))).)).))).))))))).).))))).).)....(-(((......)))-----)..........(((((.....))))) ( -27.60, z-score = -1.45, R) >droSec1.super_1 11168786 107 + 14215200 GACUAGAUGCAGCAUAGCGCUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAGGU-----GCUAUACGAUCGAUCGUAGAUGGCCG ....((((((.(((((((...(((.((((....)))).)).)...))))))).).))))).((((...(-(((......)))-----)...(((((....)))))..)))).. ( -26.80, z-score = -0.58, R) >droSim1.chr2R 12409870 107 + 19596830 GACUAGAUGCAGCAUAGCGCUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACGGAGGU-----GCUAUACGAUCGAUCGUAGAUGGCCG ....((((((.(((((((...(((.((((....)))).)).)...))))))).).))))).((((...(-(((......)))-----)...(((((....)))))..)))).. ( -26.80, z-score = -0.58, R) >droAna3.scaffold_13266 9368733 108 - 19884421 GACUAGAUGCAGCAUAGCGCUCUCAAAAUUCGUUUUCCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU-AACAACAGAUUCUU--GGCUAUAUGAUCGCUCCAAUCUGCU-- (.(.((((((.(((((((............((((....))))...))))))).).))))).).).....-.....((((((...--(((.........)))..))))))..-- ( -22.66, z-score = -0.94, R) >dp4.chr3 16483231 101 + 19779522 GACUAGAUGCAGCAUAGCGUUCUCAAAGAUCGCUUUUCAACUUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACAGAU-------GCGGUACGAUCCGAGACGCGCG---- ................((((((((...(((((((..........)))..(((((.(((((.........-......))))-------)))))).)))).)))).)))).---- ( -25.06, z-score = -0.96, R) >droPer1.super_4 6883688 101 - 7162766 GACUAGAUGCAGCAUAGCGUUCUCAAAGAUCGCUUUUCAACUUCAGCUAUGCCGACAUCUUGCCAAUUU-ACCAACAGAU-------GCGGUACGAUCCGAGACGCGCG---- ................((((((((...(((((((..........)))..(((((.(((((.........-......))))-------)))))).)))).)))).)))).---- ( -25.06, z-score = -0.96, R) >droWil1.scaffold_180700 4557894 101 + 6630534 GACUAGAUGCAGCAUAGCGCUUUUCUAAAAAUAUU------AUCAGCUAUGCCGACAUCUUGCCAAUUCGAAAAACAGAUGAUA---AUGAUGAUGCUCAAAAAAAAAAG--- .....((.((((((((((.................------....)))))))...(((((((.((..((........)))).))---).)))).)))))...........--- ( -17.00, z-score = -0.61, R) >droVir3.scaffold_12875 16937678 104 + 20611582 GACUAGAUGCAGCAUAGCGCUAUAAAAAUGCGCUUUCAAACGUCAGCUAUGCCGACAUCUUGCCAAUUG-AACAACAGAUGCGA--GGCGACGGGAUAAACAACGCG------ .....((((......(((((.........)))))......))))...(((.(((.(..(((((...(((-.....)))..))))--)..).))).))).........------ ( -24.00, z-score = -0.09, R) >droMoj3.scaffold_6496 15801376 110 - 26866924 GACUAGAUGCAGCAUAGCGCUAUGCAA-UGCGCUUUGAAACGUCAGCUAUGCCGACAUCUUGCCAAUUC-AACAAUAGAUGCGAUAAACCGAAAUAUUUAU-AAAUAUAGAUG .....((((...((.(((((.......-.))))).))...))))..(((((.((.(((((.........-......)))))))(((((........)))))-...)))))... ( -21.26, z-score = -0.18, R) >droGri2.scaffold_15245 16476686 100 + 18325388 GACUAGAUGCAGCAUAGCGCUAUAAGA-UGCGCUUUCAAACGUCAGCUAUGCCGACAUCUUGCCAAUUC-ACCAACAGAUGAGA--AUCGC---GAGAGAUAACGAG------ (.(.((((((.(((((((((.......-.))((........))..))))))).).))))).).)..(((-(.(....).)))).--.((..---..)).........------ ( -20.40, z-score = 0.25, R) >consensus GACUAGAUGCAGCAUAGCGCUCUUCAAAAUCGCUUUUCAACGUCAGCUAUGCCGACAUCUUGCCAAUUU_ACCAACAGAUGU_____GCUAUACGAUCGAUCACACAUGG___ (.(.((((((.(((((((...........................))))))).).))))).).)................................................. (-15.13 = -15.13 + -0.00)


| Location | 13,671,313 – 13,671,420 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Shannon entropy | 0.56165 |
| G+C content | 0.45961 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.81 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13671313 107 - 21146708 CGUCCAUGUCGGAUCGAUCGUAUAUC-----AACUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUC (((((.....))).))......((((-----(((...))))))-)....(((.(((((.(((((((((...((((....)))).....(.....))))))))))))))).))) ( -33.60, z-score = -1.77, R) >droEre2.scaffold_4845 7899210 107 - 22589142 GAGCCAUGUCGGAUCGAUCGUAUAGC-----ACCUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGCUUUUAAAGAGCGCUAUGCUGCAUCUAGUC ..((((...((((.............-----...)))).))))-.....(((.(((((.(((((((((....(((....)))((((.....)))))))))))))))))).))) ( -37.99, z-score = -2.56, R) >droYak2.chr2R 5627118 107 + 21139217 GAGCCAUGUCGGCUCGAUCGUAUAGC-----ACCUCUGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAUCGCUAUGCUGCAUCUAGUC (((((.....)))))...........-----(((......)))-.....(((.(((((.(((((((((....(((....)))((((.....)))))))))))))))))).))) ( -39.60, z-score = -3.13, R) >droSec1.super_1 11168786 107 - 14215200 CGGCCAUCUACGAUCGAUCGUAUAGC-----ACCUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUC ..((((..(((((....)))))....-----(((......)))-....)))).(((((.(((((((((...((((....)))).....(.....))))))))))))))).... ( -35.40, z-score = -1.91, R) >droSim1.chr2R 12409870 107 - 19596830 CGGCCAUCUACGAUCGAUCGUAUAGC-----ACCUCCGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUC ..((((..(((((....)))))....-----(((......)))-....)))).(((((.(((((((((...((((....)))).....(.....))))))))))))))).... ( -35.40, z-score = -1.91, R) >droAna3.scaffold_13266 9368733 108 + 19884421 --AGCAGAUUGGAGCGAUCAUAUAGCC--AAGAAUCUGUUGUU-AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGGAAAACGAAUUUUGAGAGCGCUAUGCUGCAUCUAGUC --((((((((.(.((.........)))--...))))))))...-.....(((.(((((.(((((((((...((((....)))).....(....).)))))))))))))).))) ( -34.60, z-score = -2.36, R) >dp4.chr3 16483231 101 - 19779522 ----CGCGCGUCUCGGAUCGUACCGC-------AUCUGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGAAGUUGAAAAGCGAUCUUUGAGAACGCUAUGCUGCAUCUAGUC ----.((((((((((((((((.((((-------((((((..(.-...)..)).))))).)))..((((....))))....)))))))..))).))))...))........... ( -31.40, z-score = -0.85, R) >droPer1.super_4 6883688 101 + 7162766 ----CGCGCGUCUCGGAUCGUACCGC-------AUCUGUUGGU-AAAUUGGCAAGAUGUCGGCAUAGCUGAAGUUGAAAAGCGAUCUUUGAGAACGCUAUGCUGCAUCUAGUC ----.((((((((((((((((.((((-------((((((..(.-...)..)).))))).)))..((((....))))....)))))))..))).))))...))........... ( -31.40, z-score = -0.85, R) >droWil1.scaffold_180700 4557894 101 - 6630534 ---CUUUUUUUUUUGAGCAUCAUCAU---UAUCAUCUGUUUUUCGAAUUGGCAAGAUGUCGGCAUAGCUGAU------AAUAUUUUUAGAAAAGCGCUAUGCUGCAUCUAGUC ---........((((((...((....---.......))...))))))..(((.(((((.(((((((((....------.................)))))))))))))).))) ( -21.90, z-score = -0.68, R) >droVir3.scaffold_12875 16937678 104 - 20611582 ------CGCGUUGUUUAUCCCGUCGCC--UCGCAUCUGUUGUU-CAAUUGGCAAGAUGUCGGCAUAGCUGACGUUUGAAAGCGCAUUUUUAUAGCGCUAUGCUGCAUCUAGUC ------.(((..((..........)).--.)))..........-.....(((.(((((.(((((((((....(((....)))((.........)))))))))))))))).))) ( -28.00, z-score = -0.15, R) >droMoj3.scaffold_6496 15801376 110 + 26866924 CAUCUAUAUUU-AUAAAUAUUUCGGUUUAUCGCAUCUAUUGUU-GAAUUGGCAAGAUGUCGGCAUAGCUGACGUUUCAAAGCGCA-UUGCAUAGCGCUAUGCUGCAUCUAGUC ...........-..........(((((((..(((.....))))-))))))((.(((((.(((((((((((.((((....))))))-..((...)))))))))))))))).)). ( -28.20, z-score = -0.96, R) >droGri2.scaffold_15245 16476686 100 - 18325388 ------CUCGUUAUCUCUC---GCGAU--UCUCAUCUGUUGGU-GAAUUGGCAAGAUGUCGGCAUAGCUGACGUUUGAAAGCGCA-UCUUAUAGCGCUAUGCUGCAUCUAGUC ------.((((........---)))).--..(((((....)))-))...(((.(((((.(((((((((....(((....)))((.-.......)))))))))))))))).))) ( -31.30, z-score = -1.57, R) >consensus ___CCAUGUGUGAUCGAUCGUAUAGC_____ACAUCUGUUGGU_AAAUUGGCAAGAUGUCGGCAUAGCUGACGUUGAAAAGCGAUUUUGAAGAGCGCUAUGCUGCAUCUAGUC .................................................(((.(((((.(((((((((....(((....)))(...........))))))))))))))).))) (-21.54 = -21.81 + 0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:37 2011