| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,661,851 – 13,661,942 |
| Length | 91 |
| Max. P | 0.819616 |

| Location | 13,661,851 – 13,661,942 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 57.98 |
| Shannon entropy | 0.89352 |
| G+C content | 0.60676 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -12.18 |
| Energy contribution | -13.83 |
| Covariance contribution | 1.65 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13661851 91 - 21146708 AGUGCGGCGAGCUGCGCUUAAAUGGCGCCAUUUGCGGUUCGG-UGGCAACGGAGGGAC---UGGAC--UACUGGACUAGUGGUCAGCCACUGGCCAA- (((((((....)))))))....(((((((......))).(((-((((..(((.....)---))(((--(((((...)))))))).))))))))))).- ( -40.10, z-score = -2.23, R) >droEre2.scaffold_4845 7890827 83 - 22589142 GGUACGGCGAGCUGCGCUUAAAUGGCGCCAUUUGCGGUUCGG-UGGCUUCGGAGG-------------GAAUGGACUAGUGGUCAGCCACUGGCCAA- (((((.(((((..(((((.....)))))..))))).)).(((-(((((((.....-------------))...(((.....))))))))))))))..- ( -33.00, z-score = -1.19, R) >droYak2.chr2R 5619552 83 + 21139217 GGUGCGGCGAGCUGCGCUUAAAUGGCGCCAUUUGCGGUUCGG-UGGCUUCGGCGG-------------GACUGGAGCUGUGGUCAGCCACUGGGCAA- ......(((((..(((((.....)))))..))))).((((((-(((((.(.((((-------------........)))).)..)))))))))))..- ( -36.20, z-score = -0.83, R) >droSec1.super_1 11160481 91 - 14215200 GGUGCGGCGAGCUGCGCUUAAAUGGCGCCAUUUGCGGUUCGG-UGGUAUCGGAGGGAC---UGGAC--UAGUGGACUAGUGGUCAGCCGCUGGCCAA- (((.(((((.((((.(((.....)))((((((...((((((.-((((.((((.....)---)))))--)).))))))))))))))))))))))))..- ( -38.00, z-score = -1.33, R) >droSim1.chr2R 12401555 93 - 19596830 GGUGCGGCGAGCUGCGCUUAAAUGGCGCCAUUUGCGGUUCGG-UGGCUUCGGAGGGAC---UGGACUAGAGUGGACUAGUGGACAGCCACUGGCCAA- (((((((....)))))))....((((.((((((..(((((((-(..(....)....))---)))))).))))))..((((((....)))))))))).- ( -39.90, z-score = -2.01, R) >droGri2.scaffold_15245 16468470 72 - 18325388 ----------------------UGUUUGCGCUCGCGCAGCGCAUGCUUUUAAGUAACGC--GUCUUCCUAAUGUGCGACCCCUUGGACAUGGACAG-- ----------------------....((((....))))(((..((((....)))).)))--((((.....((((.(((....))).))))))))..-- ( -17.70, z-score = 0.57, R) >droWil1.scaffold_181141 725006 88 + 5303230 ---GCGGUGAUUCGCGGCAAGAACUAAGCUUUAAUGGCUUC--UGGUGGCAAACCG-----AGGUUGACGGUCGUCUGCCUGCCUGCCACUGGCAGCA ---((((....))))(((.........)))......(((.(--..((((((...(.-----((((.(((....))).)))))..))))))..).))). ( -31.50, z-score = -0.33, R) >dp4.chr3 16474647 96 - 19779522 GGUUCGACGAGCUGCGCUUGAAUGGCGGCAUUUGCGGUUCAGCUCGGUUCGGCAUGGUUA-GGGCAGCCGCUGUGGCUGUGGUCUGCCACUGGUCGC- .......(((((((.(((.((((.(((.....))).))))))).)))))))....(..((-((((((((((.......)))).))))).)))..)..- ( -42.10, z-score = -1.22, R) >droPer1.super_4 6875101 97 + 7162766 GGUUCGACGAGCUGCGCUUGAAUGGCGGCAUUUGCGGUUCAGCUCGGUUCGGCAUGGUUAGGGGUAGCAGCUGUGGCUGUGGUCUGCCACUGGUCGC- ....((((((((((.(((.((((.(((.....))).))))))).))))))..((((((...(((..(((((....)))))..))))))).)))))).- ( -37.70, z-score = -0.43, R) >consensus GGUGCGGCGAGCUGCGCUUAAAUGGCGCCAUUUGCGGUUCGG_UGGCUUCGGAGGG_____GGGAC__GACUGGACUAGUGGUCAGCCACUGGCCAA_ .....((((((((((....(((((....))))))))))))......................................((((....))))..)))... (-12.18 = -13.83 + 1.65)

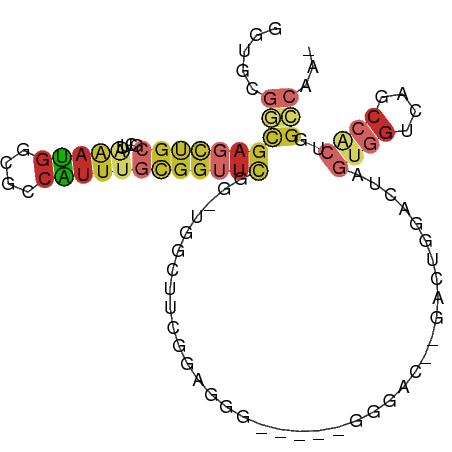

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:32 2011