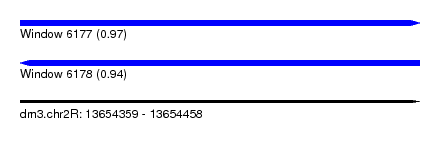
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,654,359 – 13,654,458 |
| Length | 99 |
| Max. P | 0.974251 |

| Location | 13,654,359 – 13,654,458 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.60895 |
| G+C content | 0.33257 |
| Mean single sequence MFE | -17.17 |
| Consensus MFE | -8.62 |
| Energy contribution | -8.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
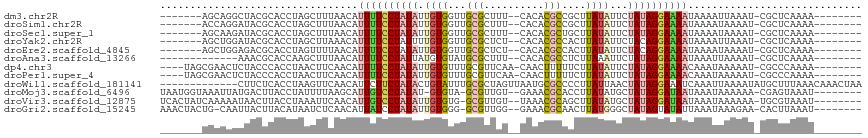
>dm3.chr2R 13654359 99 + 21146708 -------AGCAGGCUACGCACCUAGCUUUAACAUUUUCCUAUAUUGUGGUUGCGCUUU--CACACGCCGCUUAUAUUCUAUAGGAAAAUAAAAUUAAAU-CGCUCAAAA-------- -------(((((((((......))))))....(((((((((((..(((...((((...--.....).)))...)))..)))))))))))..........-.))).....-------- ( -23.60, z-score = -2.58, R) >droSim1.chr2R 12394176 99 + 19596830 -------ACCAGGAUACGCACCUAGCUUUAACAUUUUCCUAUAUUGUGGUUGCGCUUU--CACACGCCGCUUAUAUUCUAUAGGAAAAUAAAAUAAAAU-CGCUCAAAA-------- -------.....(((..((.....))......(((((((((((..(((...((((...--.....).)))...)))..)))))))))))........))-)........-------- ( -18.80, z-score = -1.64, R) >droSec1.super_1 11153066 99 + 14215200 -------AGCAAGAUACGCACCUAGCUUUAACAUUUUCCUAUAUUGUGGUUGCGCUUU--CACACGCUGCUUAUAUUCUAUAGGAAAAUAAAAUAAAAU-CGCUCAAAA-------- -------(((.......((.....))......(((((((((((..(((...((((...--.....)).))...)))..)))))))))))..........-.))).....-------- ( -20.20, z-score = -1.84, R) >droYak2.chr2R 5611924 99 - 21139217 -------AGCUGGAUACGCACCUAGCUUAAACAUUUUCCUAUUUUGUGGUUGCGCUCU--CACACGCCACUUAUAUUCUACAGGAAAAUAAAAUUAAAU-CGCUCAAAA-------- -------((((((........)))))).....((((((((....((((((.(((....--....))).)).))))......))))))))..........-.........-------- ( -18.20, z-score = -1.60, R) >droEre2.scaffold_4845 7883493 99 + 22589142 -------AGCUGGAGACGCACCUAGUUUUAACAUUUUCCUAUAUUGUGGUUGCGCUCU--CACACGCCACUUAUAUUCUACAGGAAAAUAAAAUAAAAU-CGCUCAAAA-------- -------(((..(((((.......)))))...((((((((((((.(((((((......--..)).)))))..)))).....))))))))..........-.))).....-------- ( -17.40, z-score = -0.91, R) >droAna3.scaffold_13266 9352821 93 - 19884421 -------------AAACGCACCAAGCUUUAACAUUUUCCUAUUAUGUGUAUGCGCUUU--CACACGCCUCUUAAAUUCUAUAGGAAAAUAAAUUAAAAU-CGCUCAAAA-------- -------------..........((((((((.((((((((((...((.((.(((....--....)))....)).))...))))))))))...)))))..-.))).....-------- ( -12.60, z-score = -1.40, R) >dp4.chr3 16467409 103 + 19779522 ----UAGCGAACUCUACCCACCUAACUUCAACAUUUUCCUAUAUUGUGUUUGCGUUCAA-CAACUUUUUCUUAUAUUCUAUAGGAAAACAAAUAAAAAU-CGCCCAAAA-------- ----..((((....((......)).........((((((((((..((((..........-............))))..))))))))))..........)-)))......-------- ( -11.85, z-score = -1.86, R) >droPer1.super_4 6867886 103 - 7162766 ----UAGCGAACUCUACCCACCUAACUUCAACAUUUUCCUAUAUUGUGUUUGCGUUCAA-CAACUUUUUCUUAUAUUCUAUAGGAAAACAAAUAAAAAU-CGCCCAAAA-------- ----..((((....((......)).........((((((((((..((((..........-............))))..))))))))))..........)-)))......-------- ( -11.85, z-score = -1.86, R) >droWil1.scaffold_181141 693783 104 - 5303230 -------------CUUCUCACCUAAGUUCAACAUUCUUCUAUACUGUAUUUGCGCUAGUUAAUGCGCCCCUUAUUAACUAUAGGAAAUCAAAUUAAAAUAUGCUUUAAACAAACUAA -------------...........((((........(((((((..(((...((((........))))....)))....))))))).......(((((......)))))...)))).. ( -11.60, z-score = -0.67, R) >droMoj3.scaffold_6496 15783260 104 - 26866924 UAAUGGUAAAUUAUGACUUACCUAUUUUAAGCAUUGUCCUAUAU-GUGUA-GCGUUGU--GAAACGCACCUUAUAUGCUAUAGGAUAAUAAAUAAAAAA-CGAGUAAAU-------- ....(((((........)))))..(((((...((((((((((((-(((((-(((((..--..)))))....)))))).)))))))))))...)))))..-.........-------- ( -23.90, z-score = -3.03, R) >droVir3.scaffold_12875 16920401 105 + 20611582 UCACUAUCAAAAAUAACUUACCUAAAUUCAACAUUGUCCUAUAUUGUGUG-GCGUUGU--UAAACGCAGCUUAUAUGCUAUAGGAUAAUAAAUAAAAAA-UGCGUAAAU-------- ................................(((((((((((..(((((-(.(((((--.....)))))))))))..)))))))))))..........-.........-------- ( -20.30, z-score = -2.31, R) >droGri2.scaffold_15245 16459730 104 + 18325388 AAACUACUG-CAAUUACUUACAUAAUCUCAACAUUAUCCUAUAUUGUGGG-GCGUUGG--GAAACGCAACUUAUGGGCUAUAGUAUAUUAAAUAAAGAA-CACUUAAAU-------- ..((((..(-(.........(((((............(((((...)))))-(((((..--..)))))...))))).))..))))...............-.........-------- ( -15.70, z-score = -0.64, R) >consensus _______AGCAGGAUACGCACCUAGCUUCAACAUUUUCCUAUAUUGUGGUUGCGCUCU__CACACGCCGCUUAUAUUCUAUAGGAAAAUAAAUUAAAAU_CGCUCAAAA________ .................................((((((((((.((((...(((..........)))....))))...))))))))))............................. ( -8.62 = -8.88 + 0.25)

| Location | 13,654,359 – 13,654,458 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.60895 |
| G+C content | 0.33257 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -11.77 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
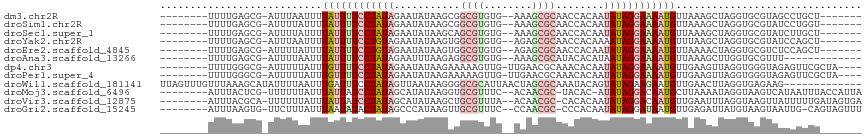
>dm3.chr2R 13654359 99 - 21146708 --------UUUUGAGCG-AUUUAAUUUUAUUUUCCUAUAGAAUAUAAGCGGCGUGUG--AAAGCGCAACCACAAUAUAGGAAAAUGUUAAAGCUAGGUGCGUAGCCUGCU------- --------.........-.((((((...(((((((((((........(..((((...--...))))..).....)))))))))))))))))((.((((.....)))))).------- ( -23.82, z-score = -0.89, R) >droSim1.chr2R 12394176 99 - 19596830 --------UUUUGAGCG-AUUUUAUUUUAUUUUCCUAUAGAAUAUAAGCGGCGUGUG--AAAGCGCAACCACAAUAUAGGAAAAUGUUAAAGCUAGGUGCGUAUCCUGGU------- --------.........-..((((...((((((((((((........(..((((...--...))))..).....)))))))))))).))))((((((.......))))))------- ( -24.12, z-score = -1.37, R) >droSec1.super_1 11153066 99 - 14215200 --------UUUUGAGCG-AUUUUAUUUUAUUUUCCUAUAGAAUAUAAGCAGCGUGUG--AAAGCGCAACCACAAUAUAGGAAAAUGUUAAAGCUAGGUGCGUAUCUUGCU------- --------.....((((-(.((((...((((((((((((...........((((...--...))))........)))))))))))).))))((.....)).....)))))------- ( -22.01, z-score = -0.86, R) >droYak2.chr2R 5611924 99 + 21139217 --------UUUUGAGCG-AUUUAAUUUUAUUUUCCUGUAGAAUAUAAGUGGCGUGUG--AGAGCGCAACCACAAAAUAGGAAAAUGUUUAAGCUAGGUGCGUAUCCAGCU------- --------.....(((.-..((((...((((((((((((.....)..((((.((((.--...))))..))))...))))))))))).))))((.....)).......)))------- ( -23.40, z-score = -1.20, R) >droEre2.scaffold_4845 7883493 99 - 22589142 --------UUUUGAGCG-AUUUUAUUUUAUUUUCCUGUAGAAUAUAAGUGGCGUGUG--AGAGCGCAACCACAAUAUAGGAAAAUGUUAAAACUAGGUGCGUCUCCAGCU------- --------.....(((.-.(((((...((((((((((((........((((.((((.--...))))..))))..)))))))))))).)))))...((.......)).)))------- ( -25.70, z-score = -2.05, R) >droAna3.scaffold_13266 9352821 93 + 19884421 --------UUUUGAGCG-AUUUUAAUUUAUUUUCCUAUAGAAUUUAAGAGGCGUGUG--AAAGCGCAUACACAUAAUAGGAAAAUGUUAAAGCUUGGUGCGUUU------------- --------..(..(((.-..((((((..((((((((((............((((...--...)))).........)))))))))))))))))))..).......------------- ( -19.90, z-score = -1.23, R) >dp4.chr3 16467409 103 - 19779522 --------UUUUGGGCG-AUUUUUAUUUGUUUUCCUAUAGAAUAUAAGAAAAAGUUG-UUGAACGCAAACACAAUAUAGGAAAAUGUUGAAGUUAGGUGGGUAGAGUUCGCUA---- --------.....((((-(.(((((((..(((..((...................((-((.......))))((((((......)))))).))..)))..))))))).))))).---- ( -20.60, z-score = -1.23, R) >droPer1.super_4 6867886 103 + 7162766 --------UUUUGGGCG-AUUUUUAUUUGUUUUCCUAUAGAAUAUAAGAAAAAGUUG-UUGAACGCAAACACAAUAUAGGAAAAUGUUGAAGUUAGGUGGGUAGAGUUCGCUA---- --------.....((((-(.(((((((..(((..((...................((-((.......))))((((((......)))))).))..)))..))))))).))))).---- ( -20.60, z-score = -1.23, R) >droWil1.scaffold_181141 693783 104 + 5303230 UUAGUUUGUUUAAAGCAUAUUUUAAUUUGAUUUCCUAUAGUUAAUAAGGGGCGCAUUAACUAGCGCAAAUACAGUAUAGAAGAAUGUUGAACUUAGGUGAGAAG------------- (((.((.(((((..((((.(((((..(((....(((.((.....)).)))((((........)))).....)))..)))))..)))))))))..)).)))....------------- ( -16.20, z-score = 0.43, R) >droMoj3.scaffold_6496 15783260 104 + 26866924 --------AUUUACUCG-UUUUUUAUUUAUUAUCCUAUAGCAUAUAAGGUGCGUUUC--ACAACGC-UACAC-AUAUAGGACAAUGCUUAAAAUAGGUAAGUCAUAAUUUACCAUUA --------.........-..(((((..((((.(((((((.........(((((((..--..)))))-.))..-.))))))).))))..)))))..(((((((....))))))).... ( -21.60, z-score = -3.33, R) >droVir3.scaffold_12875 16920401 105 - 20611582 --------AUUUACGCA-UUUUUUAUUUAUUAUCCUAUAGCAUAUAAGCUGCGUUUA--ACAACGC-CACACAAUAUAGGACAAUGUUGAAUUUAGGUAAGUUAUUUUUGAUAGUGA --------.....(((.-..............(((((((((......)).(((((..--..)))))-.......)))))))...(((..((..(((.....)))..))..)))))). ( -18.50, z-score = -1.03, R) >droGri2.scaffold_15245 16459730 104 - 18325388 --------AUUUAAGUG-UUCUUUAUUUAAUAUACUAUAGCCCAUAAGUUGCGUUUC--CCAACGC-CCCACAAUAUAGGAUAAUGUUGAGAUUAUGUAAGUAAUUG-CAGUAGUUU --------..(((((((-.....)))))))...(((((.((..((.(.((((((.((--.(((((.-.((........))....))))).))..)))))).).)).)-).))))).. ( -16.50, z-score = -0.72, R) >consensus ________UUUUGAGCG_AUUUUAAUUUAUUUUCCUAUAGAAUAUAAGCGGCGUGUG__AAAGCGCAAACACAAUAUAGGAAAAUGUUAAAGCUAGGUGCGUAUCCUGCU_______ ...........................((((((((((((...........((((........))))........))))))))))))............................... (-11.77 = -12.38 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:31 2011