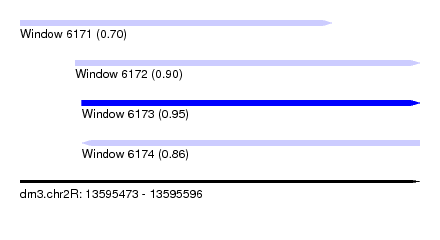
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,595,473 – 13,595,596 |
| Length | 123 |
| Max. P | 0.953341 |

| Location | 13,595,473 – 13,595,569 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.50 |
| Shannon entropy | 0.34486 |
| G+C content | 0.57263 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -24.59 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
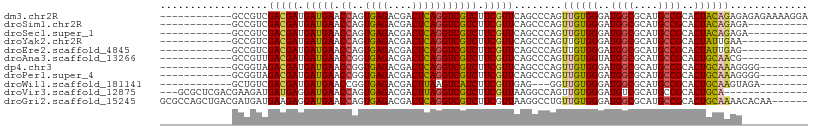
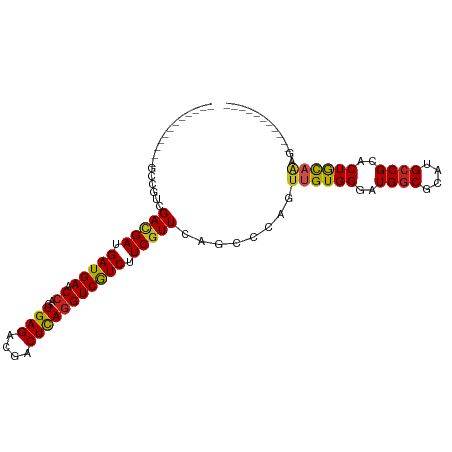
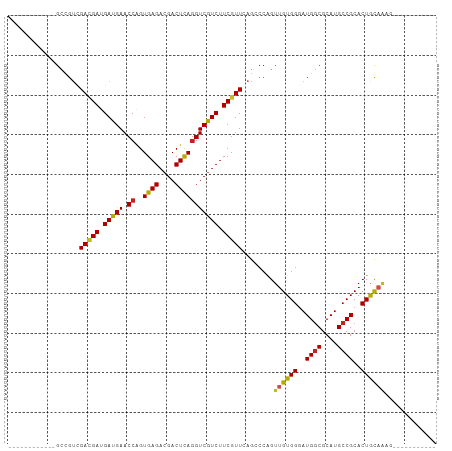
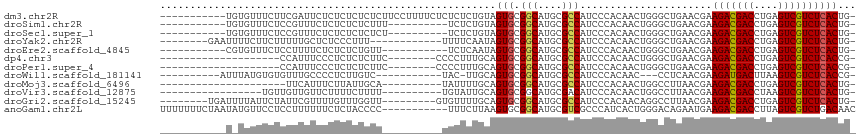
>dm3.chr2R 13595473 96 + 21146708 ------------GCCGUCGACGAUGAUGAACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGAGAGAAAAGGA ------------.((.(((((.....(((((....((((((((....)))))))).))))).....)))((((..((((....))))..)))).......))...)). ( -31.60, z-score = -0.81, R) >droSim1.chr2R 12333830 86 + 19596830 ------------GCCGUCGACGAUGAUGAACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGA---------- ------------((((((.(((((..(((((....((((((((....)))))))).))))).....))))).).)))))((.....))..........---------- ( -30.30, z-score = -0.82, R) >droSec1.super_1 11094404 86 + 14215200 ------------GCCGUCGACGAUGAUGAACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGA---------- ------------((((((.(((((..(((((....((((((((....)))))))).))))).....))))).).)))))((.....))..........---------- ( -30.30, z-score = -0.82, R) >droYak2.chr2R 5553342 85 - 21139217 ------------GCCGUCGACGAUGAUGAACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUAUUGAA----------- ------------((((((.(((((..(((((....((((((((....)))))))).))))).....))))).).)))))((.....)).........----------- ( -30.30, z-score = -1.19, R) >droEre2.scaffold_4845 7824132 85 + 22589142 ------------GCCGUCGACGAUGAUGAACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUAUUGAG----------- ------------((((((.(((((..(((((....((((((((....)))))))).))))).....))))).).)))))((.....)).........----------- ( -30.30, z-score = -1.16, R) >droAna3.scaffold_13266 9293903 85 - 19884421 ------------GCCGUUGACGAUGAUGAACCGGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGUAUGGCGCAUGCCGCACUGCAACG----------- ------------...((((((((.(((((.((..((((....))))))))))).))).)))))...((((..((.((((....)))).))..)))).----------- ( -34.50, z-score = -2.11, R) >dp4.chr3 9164865 88 - 19779522 ------------GCGGUAGACGAUGAUGAACCGGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGGGG-------- ------------((((.(((((((..(((..((......))..))).)))))))))))....(((..(((..(..((((....))))..)..)))..)))-------- ( -34.40, z-score = -1.39, R) >droPer1.super_4 1219382 88 - 7162766 ------------GCGGUAGACGAUGAUGAACCGGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGGGG-------- ------------((((.(((((((..(((..((......))..))).)))))))))))....(((..(((..(..((((....))))..)..)))..)))-------- ( -34.40, z-score = -1.39, R) >droWil1.scaffold_181141 3885334 85 + 5303230 ------------GCUGUCGACGAUGAUGAACCGGUGAGACGACUUAAGUCAUCUUCGUUGAG---GGUUGUGGGAUGGCGCAUGCCGCACUGCAAGUAGA-------- ------------(((.(((((((.(((((.....((((....))))..))))).))))))).---)))((..(..((((....))))..)..))......-------- ( -31.30, z-score = -1.97, R) >droVir3.scaffold_12875 52099 91 - 20611582 ---GCGCUCGACGAAGAUGAUGAGGAUGAACCAGUGAGACGACUUAGGUCGUCUUCGUUAAGGCCAGUUGUGGGAUGUCGCAUGCCGCACUGCA-------------- ---((.((..(((((((((((..((.....))..((((....)))).)))))))))))..))))((((.((((.(((...))).))))))))..-------------- ( -33.50, z-score = -1.64, R) >droGri2.scaffold_15245 10990867 102 + 18325388 GCGCCAGCUGACGAUGAUGAAGAGGAUGAACCAGUGAGACGACUCAGGUCGUCUUCGUUAAGGCCUGUUGUGGGAUGGCGCAUGCCGCACUGCAAAACACAA------ ((((((.((.(((((......((((((((.((..((((....))))))))))))))((....))..))))).)).)))))).(((......)))........------ ( -35.00, z-score = -1.15, R) >consensus ____________GCCGUCGACGAUGAUGAACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAG___________ ..................(((((.(((((.((..((((....))))))))))).)))))........((((((..((((....))))..))))))............. (-24.59 = -24.23 + -0.36)
| Location | 13,595,490 – 13,595,596 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.14 |
| Shannon entropy | 0.57083 |
| G+C content | 0.51556 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
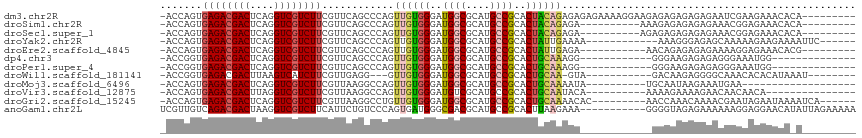
>dm3.chr2R 13595490 106 + 21146708 -ACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGAGAGAAAAGGAAGAGAGAGAGAGAAUCGAAGAAACACA--------- -....((((((((((....)))))))((.(((..((...((((((..((((....))))..))))))..........))..))).))................))).--------- ( -27.12, z-score = -0.21, R) >droSim1.chr2R 12333847 96 + 19596830 -ACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGA----------AAAGAGAGAGAGAAACGGAGAAACACA--------- -....((((((((((....)))))))((.(((.......((((((..((((....))))..))))))...----------...))).))..............))).--------- ( -26.72, z-score = -0.47, R) >droSec1.super_1 11094421 96 + 14215200 -ACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGA----------AGAGAGAGAGAGAAACGGAGAAACACA--------- -....((((((((((....)))))))((.(((.......((((((..((((....))))..))))))...----------...))).))..............))).--------- ( -26.72, z-score = -0.27, R) >droYak2.chr2R 5553359 97 - 21139217 -ACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUAUUGAAAA------------AAAGGGAGAGCAAAAAGAAGAAAAUUC------ -......((((((((....)))))))).((((..((((((.((((.(((...))).))))))).((....)------------)..))).))))................------ ( -27.90, z-score = -1.44, R) >droEre2.scaffold_4845 7824149 95 + 22589142 -ACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUAUUGAGA-----------AACAGAGAGAGAAAAGGAGAAACACG--------- -....((((((((((....)))))))((.(((..((((....))))..(((....)))..........))-----------)...))................))).--------- ( -23.90, z-score = 0.30, R) >dp4.chr3 9164882 89 - 19779522 -ACCGGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGG------------GGGAAGAGAGAGGGAAAUGG-------------- -.((....(((((((....)))))))((.(((..(((..(((..(..((((....))))..)..)))...------------)))..))).)))).......-------------- ( -33.60, z-score = -1.55, R) >droPer1.super_4 1219399 89 - 7162766 -ACCGGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGG------------GGGAAGAGAGAGGGAAAUGG-------------- -.((....(((((((....)))))))((.(((..(((..(((..(..((((....))))..)..)))...------------)))..))).)))).......-------------- ( -33.60, z-score = -1.55, R) >droWil1.scaffold_181141 3885351 92 + 5303230 -ACCGGUGAGACGACUUAAGUCAUCUUCGUUGAGG---GUUGUGGGAUGGCGCAUGCCGCACUGCAA-GUA-----------GACAAGAGGGGCAAACACACAUAAAU-------- -....(((.(((.......))).(((((.(((..(---.(((..(..((((....))))..)..)))-.).-----------..)))))))).....)))........-------- ( -23.90, z-score = -0.13, R) >droMoj3.scaffold_6496 5562221 86 + 26866924 -ACCAGUGAGACGACUCAGGUCGUCUUCGUUAAGGCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAAUA----------UGCAAUAAGAAAUGAA------------------- -......((((((((....)))))))).......((...(((..(..((((....))))..)..)))....----------.)).............------------------- ( -24.30, z-score = -0.63, R) >droVir3.scaffold_12875 52125 90 - 20611582 -ACCAGUGAGACGACUUAGGUCGUCUUCGUUAAGGCCAGUUGUGGGAUGUCGCAUGCCGCACUGCAAUACA----------AAAAGAAAAGAACAACAACA--------------- -......((((((((....)))))))).((....((.(((.((((.(((...))).)))))))))...)).----------....................--------------- ( -23.60, z-score = -0.97, R) >droGri2.scaffold_15245 10990896 100 + 18325388 -ACCAGUGAGACGACUCAGGUCGUCUUCGUUAAGGCCUGUUGUGGGAUGGCGCAUGCCGCACUGCAAAACAC---------AACCAAACAAAACGAAUAGAAUAAAAUCA------ -.....(((((((((....))))))((((((..(....((((((....(((....)))((...))....)))---------)))....)..))))))..........)))------ ( -25.50, z-score = -0.82, R) >anoGam1.chr2L 31452038 105 - 48795086 UCGUUGUCAGACGACUAAGGUCGUCUUCAUUCUGUCCCAGUGAUGGGCGACGCAUGCCGCACUUAAGAAA-----------GGGGUAGAGAAAAAAGGAGGAACAUAUUAGAAAAA ((.((.(((((((((....)))))))....(((..(((((((...((((.....)))).)))).......-----------)))..)))))...)).))................. ( -28.01, z-score = -1.18, R) >consensus _ACCAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGA____________AGAAAGAGAGAAAAAAAGAAACA_A_________ .......((((((((....))))))))............((((((..((((....))))..))))))................................................. (-19.72 = -19.70 + -0.02)

| Location | 13,595,492 – 13,595,596 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.71 |
| Shannon entropy | 0.57010 |
| G+C content | 0.51519 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
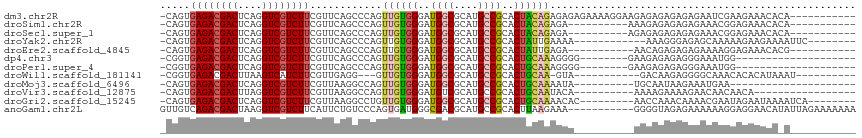
>dm3.chr2R 13595492 104 + 21146708 -CAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGAGAGAAAAGGAAGAGAGAGAGAGAAUCGAAGAAACACA----------- -..((((((((((....)))))))((.(((..((...((((((..((((....))))..))))))..........))..))).))................))).----------- ( -27.12, z-score = -0.47, R) >droSim1.chr2R 12333849 94 + 19596830 -CAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGA----------AAAGAGAGAGAGAAACGGAGAAACACA----------- -..((((((((((....)))))))((.(((.......((((((..((((....))))..))))))...----------...))).))..............))).----------- ( -26.72, z-score = -0.73, R) >droSec1.super_1 11094423 94 + 14215200 -CAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUACAGAGA----------AGAGAGAGAGAGAAACGGAGAAACACA----------- -..((((((((((....)))))))((.(((.......((((((..((((....))))..))))))...----------...))).))..............))).----------- ( -26.72, z-score = -0.56, R) >droYak2.chr2R 5553361 95 - 21139217 -CAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUAUUGAAAA------------AAAGGGAGAGCAAAAAGAAGAAAAUUC-------- -....((((((((....)))))))).((((..((((((.((((.(((...))).))))))).((....)------------)..))).))))................-------- ( -27.90, z-score = -1.77, R) >droEre2.scaffold_4845 7824151 93 + 22589142 -CAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUAUUGAGA-----------AACAGAGAGAGAAAAGGAGAAACACG----------- -..((((((((((....)))))))((.(((..((((....))))..(((....)))..........))-----------)...))................))).----------- ( -23.90, z-score = 0.05, R) >dp4.chr3 9164884 87 - 19779522 -CGGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGGGG--------GAAGAGAGAGGGAAAUGG-------------------- -.....(((((((....)))))))((.(((..(((..(((..(..((((....))))..)..)))...))--------)..))).)).........-------------------- ( -31.20, z-score = -1.41, R) >droPer1.super_4 1219401 87 - 7162766 -CGGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGGGG--------GAAGAGAGAGGGAAAUGG-------------------- -.....(((((((....)))))))((.(((..(((..(((..(..((((....))))..)..)))...))--------)..))).)).........-------------------- ( -31.20, z-score = -1.41, R) >droWil1.scaffold_181141 3885353 90 + 5303230 -CGGUGAGACGACUUAAGUCAUCUUCGUUGAGG---GUUGUGGGAUGGCGCAUGCCGCACUGCAA-GUA-----------GACAAGAGGGGCAAACACACAUAAAU---------- -..(((.(((.......))).(((((.(((..(---.(((..(..((((....))))..)..)))-.).-----------..)))))))).....)))........---------- ( -23.90, z-score = -0.67, R) >droMoj3.scaffold_6496 5562223 84 + 26866924 -CAGUGAGACGACUCAGGUCGUCUUCGUUAAGGCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAAUA----------UGCAAUAAGAAAUGAA--------------------- -....((((((((....)))))))).......((...(((..(..((((....))))..)..)))....----------.)).............--------------------- ( -24.30, z-score = -0.92, R) >droVir3.scaffold_12875 52127 88 - 20611582 -CAGUGAGACGACUUAGGUCGUCUUCGUUAAGGCCAGUUGUGGGAUGUCGCAUGCCGCACUGCAAUACA----------AAAAGAAAAGAACAACAACA----------------- -....((((((((....)))))))).((....((.(((.((((.(((...))).)))))))))...)).----------....................----------------- ( -23.60, z-score = -1.21, R) >droGri2.scaffold_15245 10990898 98 + 18325388 -CAGUGAGACGACUCAGGUCGUCUUCGUUAAGGCCUGUUGUGGGAUGGCGCAUGCCGCACUGCAAAACAC---------AACCAAACAAAACGAAUAGAAUAAAAUCA-------- -...(((((((((....))))))((((((..(....((((((....(((....)))((...))....)))---------)))....)..))))))..........)))-------- ( -25.50, z-score = -1.07, R) >anoGam1.chr2L 31452040 105 - 48795086 GUUGUCAGACGACUAAGGUCGUCUUCAUUCUGUCCCAGUGAUGGGCGACGCAUGCCGCACUUAAGAAA-----------GGGGUAGAGAAAAAAGGAGGAACAUAUUAGAAAAAAA .((.(((((((((....)))))))...((((..(((((((...((((.....)))).)))).......-----------)))..)))).......)).))................ ( -28.01, z-score = -1.55, R) >consensus _CAGUGAGACGACUCAGGUCGUCUUCGUUCAGCCCAGUUGUGGGAUGGCGCAUGCCGCACUGCAAAGAA__________AAGAGAGAGAGAAAAGGAGAAACA_A___________ .....((((((((....))))))))............((((((..((((....))))..))))))................................................... (-19.72 = -19.70 + -0.02)

| Location | 13,595,492 – 13,595,596 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.71 |
| Shannon entropy | 0.57010 |
| G+C content | 0.51519 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.06 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
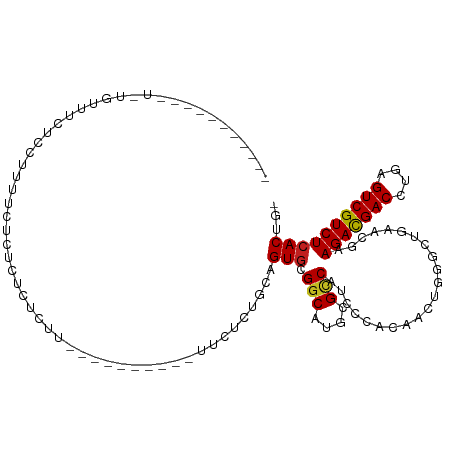
>dm3.chr2R 13595492 104 - 21146708 -----------UGUGUUUCUUCGAUUCUCUCUCUCUCUUCCUUUUCUCUCUCUGUAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACUG- -----------.(((......((((((....((..(((((...(((...(((.((.(((.(((....)))....))).)).)))..))).))))).))...))))))...)))..- ( -23.20, z-score = -0.41, R) >droSim1.chr2R 12333849 94 - 19596830 -----------UGUGUUUCUCCGUUUCUCUCUCUCUUU----------UCUCUGUAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACUG- -----------.(((......(((((............----------............(((....)))..((((....))))..))))).(((((((....))))))))))..- ( -22.40, z-score = -0.26, R) >droSec1.super_1 11094423 94 - 14215200 -----------UGUGUUUCUCCGUUUCUCUCUCUCUCU----------UCUCUGUAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACUG- -----------.(((......(((((............----------............(((....)))..((((....))))..))))).(((((((....))))))))))..- ( -22.40, z-score = -0.20, R) >droYak2.chr2R 5553361 95 + 21139217 --------GAAUUUUCUUCUUUUUGCUCUCCCUUU------------UUUUCAAUAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACUG- --------...........................------------.......(((((.(((....)))..((((....))))........(((((((....))))))))))))- ( -20.10, z-score = -0.66, R) >droEre2.scaffold_4845 7824151 93 - 22589142 -----------CGUGUUUCUCCUUUUCUCUCUCUGUU-----------UCUCAAUAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACUG- -----------.(((..................((((-----------(...........(((....)))..((((....))))..))))).(((((((....))))))))))..- ( -20.70, z-score = -0.02, R) >dp4.chr3 9164884 87 + 19779522 --------------------CCAUUUCCCUCUCUCUUC--------CCCCUUUGCAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACCG- --------------------..................--------........((((..(((....)))...(((....)))))))...((.((((((....))))))))....- ( -19.90, z-score = -0.56, R) >droPer1.super_4 1219401 87 + 7162766 --------------------CCAUUUCCCUCUCUCUUC--------CCCCUUUGCAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACCG- --------------------..................--------........((((..(((....)))...(((....)))))))...((.((((((....))))))))....- ( -19.90, z-score = -0.56, R) >droWil1.scaffold_181141 3885353 90 - 5303230 ----------AUUUAUGUGUGUUUGCCCCUCUUGUC-----------UAC-UUGCAGUGCGGCAUGCGCCAUCCCACAAC---CCUCAACGAAGAUGACUUAAGUCGUCUCACCG- ----------....(((.((((.((((.(.((.((.-----------...-..)))).).)))).)))))))........---.......((.((((((....))))))))....- ( -21.90, z-score = -2.09, R) >droMoj3.scaffold_6496 5562223 84 - 26866924 ---------------------UUCAUUUCUUAUUGCA----------UAUUUUGCAGUGCGGCAUGCGCCAUCCCACAACUGGCCUUAACGAAGACGACCUGAGUCGUCUCACUG- ---------------------..(((.((.(((((((----------.....))))))).)).))).((((.........))))......((.((((((....))))))))....- ( -22.00, z-score = -2.04, R) >droVir3.scaffold_12875 52127 88 + 20611582 -----------------UGUUGUUGUUCUUUUCUUUU----------UGUAUUGCAGUGCGGCAUGCGACAUCCCACAACUGGCCUUAACGAAGACGACCUAAGUCGUCUCACUG- -----------------..((((((...........(----------(((((.((......))))))))....(((....)))...))))))(((((((....))))))).....- ( -17.90, z-score = 0.13, R) >droGri2.scaffold_15245 10990898 98 - 18325388 --------UGAUUUUAUUCUAUUCGUUUUGUUUGGUU---------GUGUUUUGCAGUGCGGCAUGCGCCAUCCCACAACAGGCCUUAACGAAGACGACCUGAGUCGUCUCACUG- --------(((..........((((((..(((((.((---------(((....((...))(((....)))....))))))))))...))))))((((((....)))))))))...- ( -27.30, z-score = -1.58, R) >anoGam1.chr2L 31452040 105 + 48795086 UUUUUUUCUAAUAUGUUCCUCCUUUUUUCUCUACCCC-----------UUUCUUAAGUGCGGCAUGCGUCGCCCAUCACUGGGACAGAAUGAAGACGACCUUAGUCGUCUGACAAC ......((.....((((((.................(-----------((....))).(((((....)))))........))))))....))(((((((....)))))))...... ( -23.30, z-score = -1.64, R) >consensus ___________U_UGUUUCUCCUUUUCUCUCUCUCUU__________UUCUCUGCAGUGCGGCAUGCGCCAUCCCACAACUGGGCUGAACGAAGACGACCUGAGUCGUCUCACUG_ ........................................................(((.(((....)))......................(((((((....))))))))))... (-14.04 = -14.06 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:28 2011