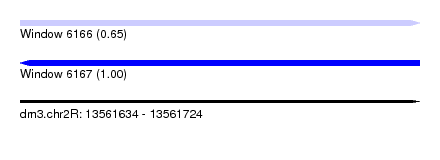
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,561,634 – 13,561,724 |
| Length | 90 |
| Max. P | 0.999387 |

| Location | 13,561,634 – 13,561,724 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 60.34 |
| Shannon entropy | 0.82165 |
| G+C content | 0.50789 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -6.32 |
| Energy contribution | -5.61 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
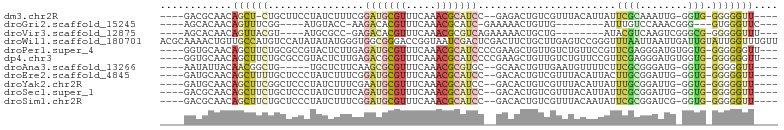
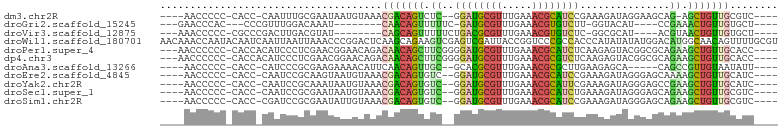
>dm3.chr2R 13561634 90 + 21146708 ----GACGCAACAGCU-CUGCUUCCUAUCUUUCGGAUGCGUUUCAAACGCAUCC--GAGACUGUCGUUUACAUUAUUCGCAAAUUG-GGUG-GGGGGUU---- ----((.((....)))-).((((((((((((((((((((((.....))))))))--)))).((.((..((...))..)))).....-))))-)))))).---- ( -31.50, z-score = -3.39, R) >droGri2.scaffold_15245 14697625 79 - 18325388 ----AGCACAACAGUUUCGG----AUGUACC-AAGACACGUUUCAAACGCAUC-GAAAAACUGUUG--------AUUUGUCCAAACGGG---GUGGGUUC--- ----....(((((((((..(----((((...-..((......))....)))))-...)))))))))--------(((..(((.....))---)..)))..--- ( -18.70, z-score = -0.26, R) >droVir3.scaffold_12875 10103717 82 - 20611582 ----AGCACAACAGUUACGU----AUGCGCC-GAGACACGUUUCAAACGCGUCAGAAAAACUGCUG--------AUACGUCAAGUCGGGCG-GGGGGUUU--- ----.(((..((......))----.)))(((-..(((.(((.....))).))).......((((((--------((.......))).))))-)..)))..--- ( -17.90, z-score = 1.17, R) >droWil1.scaffold_180701 1112998 103 - 3904529 ACGCAAAACUGUUGCCAUGUCCAUAUAUAUGGGUGGCGGGACCGGUAAUCGACUCGACUUCUGCUUGAGUCCGGGUUUAAUUAAUUGAUUGUAUUGGUUUGUU ..(((...((((..((((((......)))).))..))))((((((((...((((((((((......)))).))))))(((((....)))))))))))))))). ( -26.80, z-score = -0.48, R) >droPer1.super_4 1185299 95 - 7162766 ----GGUGCAACAGCUUCUGCGCCGUACUCUUGAGAUGCGUUUCAAACGCAUCCCCGAAGCUGUUGUCUGUUCCGUUCGAGGGAUGUGGUG-GGGGGGUU--- ----........(((((((.(((((((((((((((((((((.....)))))))..((.(((........))).)).))))))).)))))))-.)))))))--- ( -34.50, z-score = -0.95, R) >dp4.chr3 9131211 95 - 19779522 ----GGUGCAACAGCUUCUGCGCCGUACUCUUGAGACGCGUUUCAAACGCAUCCCCGAAGCUGUUGUCUGUUCCGUUCGAGGGAUGUGGUG-GGGGGGUU--- ----((((((........))))))..((((((((((....))))...((((((((((((((........))....)))).))))))))...-.)))))).--- ( -32.80, z-score = -0.17, R) >droAna3.scaffold_13266 9262863 86 - 19884421 ----AAUAUUACAACGGCUG-----UGCUCUUCAAGCGCGUUUCAAACGCGUGC--GCAACUGUUGAAUGUUUUCUUCGCGGGAUG-GGUG-GGGGGUU---- ----((((((.((((((.((-----(((.......((((((.....))))))))--))).))))))))))))((((((((......-.)))-)))))..---- ( -29.01, z-score = -2.03, R) >droEre2.scaffold_4845 7791083 91 + 22589142 ----GAUGCAACAGCUUUUGCUCCCUAUCUUUCGGAUGCGUUUCAAACGCAUCC--GACACUGUCGUUUACAUUACUUGCGGAUUG-GGUG-GGGGGUU---- ----...((....))....(((((((((((.((((((((((.....))))))))--))..((((.((.......))..))))...)-))))-)))))).---- ( -34.60, z-score = -4.00, R) >droYak2.chr2R 5517020 91 - 21139217 ----GAUGCAACAGCUUCGGCUCCCUAUCUUUCGAAUGCGUUUCAAACGCAUCC--GACACUGUCGUUUACAUUAUUUGCGGAUUG-GGUG-GGGGGUU---- ----((.((....)).))(((((((((((..(((.((((((.....)))))).)--))((...((((...........))))..))-))))-)))))))---- ( -29.60, z-score = -2.19, R) >droSec1.super_1 11060867 91 + 14215200 ----GACGCAACAGCUUCUGCUCCCUAUCUUUCAGAUGCGUUUCAAACGCAUCC--GACACUGUCGUUUACAUUAUUCGCGGAUUG-GGUG-GGGGGUU---- ----((.((....)).)).(((((((((((.((.(((((((.....)))))))(--(((...))))...............))..)-))))-)))))).---- ( -30.70, z-score = -2.79, R) >droSim1.chr2R 12298378 91 + 19596830 ----GACGCAACAGCUUCUGCUCCCUAUCUUUCGGAUGCGUUUCAAACGCAUCC--GACACUGUCGUUUACAAUAUUCGCGGAUCG-GGUG-GGGGGUU---- ----((.((....)).)).(((((((((((.((((((((((.....))))))))--))..(((.((..((...))..)))))...)-))))-)))))).---- ( -36.50, z-score = -4.25, R) >consensus ____GACGCAACAGCUUCUGCUCCCUAUCUUUCAGAUGCGUUUCAAACGCAUCC__GAAACUGUUGUUUACAUUAUUCGCGGAAUG_GGUG_GGGGGUU____ ....(((((....))...................(((((((.....))))))).........)))...................................... ( -6.32 = -5.61 + -0.71)
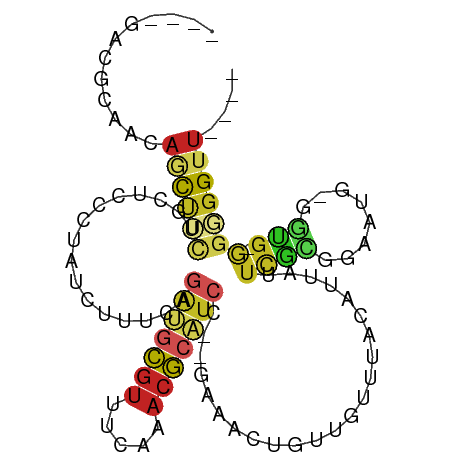
| Location | 13,561,634 – 13,561,724 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 60.34 |
| Shannon entropy | 0.82165 |
| G+C content | 0.50789 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -12.64 |
| Energy contribution | -12.34 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13561634 90 - 21146708 ----AACCCCC-CACC-CAAUUUGCGAAUAAUGUAAACGACAGUCUC--GGAUGCGUUUGAAACGCAUCCGAAAGAUAGGAAGCAG-AGCUGUUGCGUC---- ----.......-....-...((((((.....))))))((((((.(((--((((((((.....))))))))........(....).)-))))))))....---- ( -25.40, z-score = -2.77, R) >droGri2.scaffold_15245 14697625 79 + 18325388 ---GAACCCAC---CCCGUUUGGACAAAU--------CAACAGUUUUUC-GAUGCGUUUGAAACGUGUCUU-GGUACAU----CCGAAACUGUUGUGCU---- ---....(((.---......)))......--------((((((((((..-(((((((.....)))))))..-((.....----))))))))))))....---- ( -19.50, z-score = -1.12, R) >droVir3.scaffold_12875 10103717 82 + 20611582 ---AAACCCCC-CGCCCGACUUGACGUAU--------CAGCAGUUUUUCUGACGCGUUUGAAACGUGUCUC-GGCGCAU----ACGUAACUGUUGUGCU---- ---........-.((.((((...((((((--------..((((.....))(((((((.....)))))))..-.))..))----))))....)))).)).---- ( -22.90, z-score = -1.63, R) >droWil1.scaffold_180701 1112998 103 + 3904529 AACAAACCAAUACAAUCAAUUAAUUAAACCCGGACUCAAGCAGAAGUCGAGUCGAUUACCGGUCCCGCCACCCAUAUAUAUGGACAUGGCAACAGUUUUGCGU .....(((((.((................((((..((.....))(((((...))))).))))....((((.((((....))))...))))....)).))).)) ( -18.50, z-score = -0.50, R) >droPer1.super_4 1185299 95 + 7162766 ---AACCCCCC-CACCACAUCCCUCGAACGGAACAGACAACAGCUUCGGGGAUGCGUUUGAAACGCAUCUCAAGAGUACGGCGCAGAAGCUGUUGCACC---- ---........-.......(((.......)))...(.((((((((((.(((((((((.....)))))))))....((.....)).)))))))))))...---- ( -31.00, z-score = -2.63, R) >dp4.chr3 9131211 95 + 19779522 ---AACCCCCC-CACCACAUCCCUCGAACGGAACAGACAACAGCUUCGGGGAUGCGUUUGAAACGCGUCUCAAGAGUACGGCGCAGAAGCUGUUGCACC---- ---........-.......(((.......)))...(.((((((((((.(((((((((.....)))))))))....((.....)).)))))))))))...---- ( -30.60, z-score = -1.92, R) >droAna3.scaffold_13266 9262863 86 + 19884421 ----AACCCCC-CACC-CAUCCCGCGAAGAAAACAUUCAACAGUUGC--GCACGCGUUUGAAACGCGCUUGAAGAGCA-----CAGCCGUUGUAAUAUU---- ----.......-....-.................(((((((.((((.--((.(((((.....)))))(.....).)).-----)))).)))).)))...---- ( -15.60, z-score = -0.67, R) >droEre2.scaffold_4845 7791083 91 - 22589142 ----AACCCCC-CACC-CAAUCCGCAAGUAAUGUAAACGACAGUGUC--GGAUGCGUUUGAAACGCAUCCGAAAGAUAGGGAGCAAAAGCUGUUGCAUC---- ----.......-..((-(.((((....).................((--((((((((.....))))))))))..))).))).((((......))))...---- ( -27.60, z-score = -3.32, R) >droYak2.chr2R 5517020 91 + 21139217 ----AACCCCC-CACC-CAAUCCGCAAAUAAUGUAAACGACAGUGUC--GGAUGCGUUUGAAACGCAUUCGAAAGAUAGGGAGCCGAAGCUGUUGCAUC---- ----.......-....-....................(((((((.((--((((((((.....))))))((....)).......)))).)))))))....---- ( -23.80, z-score = -1.81, R) >droSec1.super_1 11060867 91 - 14215200 ----AACCCCC-CACC-CAAUCCGCGAAUAAUGUAAACGACAGUGUC--GGAUGCGUUUGAAACGCAUCUGAAAGAUAGGGAGCAGAAGCUGUUGCGUC---- ----.......-..((-(.((((((.................)))((--((((((((.....))))))))))..))).))).((((......))))...---- ( -24.53, z-score = -1.66, R) >droSim1.chr2R 12298378 91 - 19596830 ----AACCCCC-CACC-CGAUCCGCGAAUAUUGUAAACGACAGUGUC--GGAUGCGUUUGAAACGCAUCCGAAAGAUAGGGAGCAGAAGCUGUUGCGUC---- ----.......-..((-(.(((......((((((.....))))))((--((((((((.....))))))))))..))).))).((((......))))...---- ( -29.50, z-score = -2.98, R) >consensus ____AACCCCC_CACC_CAAUCCGCGAAUAAUGUAAACAACAGUUUC__GGAUGCGUUUGAAACGCAUCUGAAAGAUAGGGAGCAGAAGCUGUUGCAUC____ .....................................(((((((.((..((((((((.....))))))))...............)).)))))))........ (-12.64 = -12.34 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:22 2011