| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,556,457 – 13,556,550 |
| Length | 93 |
| Max. P | 0.999966 |

| Location | 13,556,457 – 13,556,550 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 62.30 |
| Shannon entropy | 0.74640 |
| G+C content | 0.48680 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -16.64 |
| Energy contribution | -15.39 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.64 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.35 |
| SVM RNA-class probability | 0.999966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
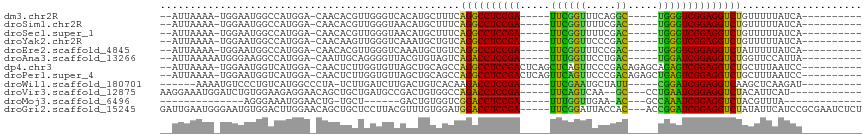
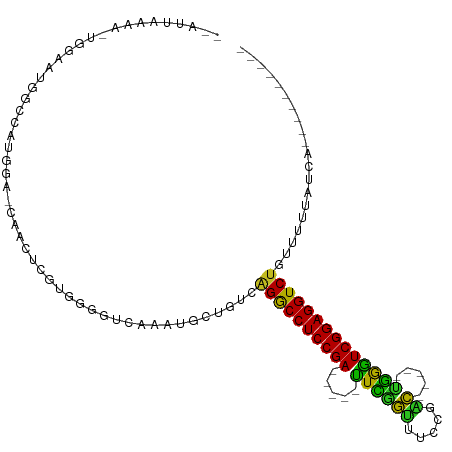
>dm3.chr2R 13556457 93 + 21146708 --AUUAAAA-UGGAAUGGCCAUGGA-CAACACGUUGGGUCACAUGCUUUCAGGCCUCCGA-----UUCGGUUUCAGGC-----UGGGUCGGAGGUCUGUUUUUAUCA---------- --......(-((((((((((...((-(.....))).)))))........(((((((((((-----((((((.....))-----))))))))))))))).))))))..---------- ( -36.40, z-score = -3.91, R) >droSim1.chr2R 12293146 93 + 19596830 --AUUAAAA-UGGAAUGGCCAUGGA-CAACACGUUGGGUAACAUGCUUUCAGGCCUCCGA-----UUCGGUUUUCGAC-----UGGGUCGGAGGUCUGUUUUUAUCA---------- --......(-(((((.(((.(((..-(((....))).....))))))..(((((((((((-----(((((((...)))-----))))))))))))))).))))))..---------- ( -33.60, z-score = -3.33, R) >droSec1.super_1 11055722 93 + 14215200 --AUUAAAA-UGGAAUGGCCAUGGA-CAACACGUUGGGUAACAUGCUUUCAGGCCUCCGA-----UUCGGUUUUCGAC-----UGGGUCGGAGGUCUGUUUUUAUCA---------- --......(-(((((.(((.(((..-(((....))).....))))))..(((((((((((-----(((((((...)))-----))))))))))))))).))))))..---------- ( -33.60, z-score = -3.33, R) >droYak2.chr2R 5511957 93 - 21139217 --AUUAAAA-UGGAAUGGCCAUGGA-CAACAAGUUGGGUCAAAUGCUGUCAGGCCUCCGA-----UUCGGUUCCCGAC-----UGGGUCGGAGGUCUGUUUUUAUCA---------- --......(-((((((((((...((-(.....))).)))))........(((((((((((-----(((((((...)))-----))))))))))))))).))))))..---------- ( -36.50, z-score = -3.26, R) >droEre2.scaffold_4845 7785949 93 + 22589142 --AUUAAAA-UGGAAUGGCCAUGGA-CAACACGUUGGGUCAAAUGCUGUCAGGCCUCCGA-----UUCGGUUUCCGAC-----UGGGUCGGAGGUCUAUUUUUAUCA---------- --......(-((((((((((...((-(.....))).))))).........((((((((((-----(((((((...)))-----))))))))))))))..))))))..---------- ( -34.20, z-score = -3.03, R) >droAna3.scaffold_13266 9258475 94 - 19884421 --AUUAAAAAUGGGAAGGCCAUGGA-CAAUUGCAGGGGUUACGUGUAGUCAGACCUCCGA-----UUUGGUUCCUGAC-----UGGAUCGGAGGUCUGGUUCCAUUA---------- --......(((((((..........-..((((((.(.....).))))))(((((((((((-----((..(((...)))-----..))))))))))))).))))))).---------- ( -41.20, z-score = -4.50, R) >dp4.chr3 9125945 103 - 19779522 --AUUAAAA-UGGAAUGGUCAUGGA-CAACUCUUGGUGUUAGCUGCAGCCAGGCCUCCGACUCAGCUCAGUUCCCGACAGAGCAGAGUCGGAGGUCUGCUUUAAUCC---------- --.......-.(((...((....((-((.(....).))))....))(((.(((((((((((((.((((.(((...))).)))).))))))))))))))))....)))---------- ( -39.60, z-score = -2.85, R) >droPer1.super_4 1179996 103 - 7162766 --AUUAAAA-UGGAAUGGUCAUGGA-CAACUCUUGGUGUUAGCUGCAGCCAGGCCUCCGACUCAGUUCAGUUCCCGACAGAGCUGAGUCGGAGGUCUGCUUUAAUCC---------- --.......-.(((...((....((-((.(....).))))....))(((.((((((((((((((((((.(((...))).)))))))))))))))))))))....)))---------- ( -42.50, z-score = -3.90, R) >droWil1.scaffold_180701 1108610 90 - 3904529 ------AAAAUGUCCCUGUCAUGGCCCUA-UCUUGAUCUUGACUGUCACAAGACCUCCGA-----UUCGAAUGCUAUU-----CGGAUCGGAGGUCAAGCUCAAGAU---------- ------.....(..((......))..).(-((((((.((((((.(((....)))((((((-----(((((((...)))-----)))))))))))))))).)))))))---------- ( -37.00, z-score = -5.43, R) >droVir3.scaffold_12875 11293791 95 + 20611582 AAGGAAAUGGAUCUGUGGAAGAGGAACAGCUGCUGAUGCCGACUGUGGCCAGACCUCCGA-----UUCAGUCAA--GC---CCUGAAUCGGAGGUCUACAUUCAU------------ ......((((((..........((.(((((.((....)).).))))..))((((((((((-----(((((....--..---.)))))))))))))))..))))))------------ ( -38.40, z-score = -3.32, R) >droMoj3.scaffold_6496 11237556 74 - 26866924 --------------AGGGAAAUGGAACUG-UGCU------GACUGUGGUCGGACCUCCGA-----UUUGGUUGAA-AC---GCCAAAUCGGAGGUCUACGUUUA------------- --------------....(((((......-....------(((....)))((((((((((-----((((((....-..---)))))))))))))))).))))).------------- ( -33.50, z-score = -5.10, R) >droGri2.scaffold_15245 4890715 109 - 18325388 GAUUGAAUGGGAAUGUGGACUUGGAACAGCUGCUCCUUACGUUUGUGGAUGGACCUCCGA-----UUCGGAUUACCAC---ACCGGAUCGGAGGUCUAUAUUCAUCCGCGAAUCUCU ........((((..(..(.((......)))..)))))...((((((((((((((((((((-----(((((........---.))))))))))))))......))))))))))).... ( -43.20, z-score = -3.78, R) >consensus __AUUAAAA_UGGAAUGGCCAUGGA_CAACUCGUGGGGUCAAAUGCUGUCAGGCCUCCGA_____UUCGGUUUCCGAC_____UGGGUCGGAGGUCUGUUUUUAUCA__________ ..................................................((((((((((.....((((..............)))))))))))))).................... (-16.64 = -15.39 + -1.25)
| Location | 13,556,457 – 13,556,550 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 62.30 |
| Shannon entropy | 0.74640 |
| G+C content | 0.48680 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -9.85 |
| Energy contribution | -9.84 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13556457 93 - 21146708 ----------UGAUAAAAACAGACCUCCGACCCA-----GCCUGAAACCGAA-----UCGGAGGCCUGAAAGCAUGUGACCCAACGUGUUG-UCCAUGGCCAUUCCA-UUUUAAU-- ----------.(((.....(((.(((((((..(.-----..........)..-----))))))).)))..(((((((......))))))))-)).((((.....)))-)......-- ( -23.00, z-score = -2.22, R) >droSim1.chr2R 12293146 93 - 19596830 ----------UGAUAAAAACAGACCUCCGACCCA-----GUCGAAAACCGAA-----UCGGAGGCCUGAAAGCAUGUUACCCAACGUGUUG-UCCAUGGCCAUUCCA-UUUUAAU-- ----------.(((.....(((.(((((((....-----.(((.....))).-----))))))).)))..((((((((....)))))))))-)).((((.....)))-)......-- ( -25.70, z-score = -3.29, R) >droSec1.super_1 11055722 93 - 14215200 ----------UGAUAAAAACAGACCUCCGACCCA-----GUCGAAAACCGAA-----UCGGAGGCCUGAAAGCAUGUUACCCAACGUGUUG-UCCAUGGCCAUUCCA-UUUUAAU-- ----------.(((.....(((.(((((((....-----.(((.....))).-----))))))).)))..((((((((....)))))))))-)).((((.....)))-)......-- ( -25.70, z-score = -3.29, R) >droYak2.chr2R 5511957 93 + 21139217 ----------UGAUAAAAACAGACCUCCGACCCA-----GUCGGGAACCGAA-----UCGGAGGCCUGACAGCAUUUGACCCAACUUGUUG-UCCAUGGCCAUUCCA-UUUUAAU-- ----------...(((((.......((((((...-----)))))).......-----..(((((((.(((((((..(......)..)))))-))...))))..))).-)))))..-- ( -26.00, z-score = -2.12, R) >droEre2.scaffold_4845 7785949 93 - 22589142 ----------UGAUAAAAAUAGACCUCCGACCCA-----GUCGGAAACCGAA-----UCGGAGGCCUGACAGCAUUUGACCCAACGUGUUG-UCCAUGGCCAUUCCA-UUUUAAU-- ----------...(((((.......((((((...-----)))))).......-----..(((((((.((((((((.(......).))))))-))...))))..))).-)))))..-- ( -28.00, z-score = -3.33, R) >droAna3.scaffold_13266 9258475 94 + 19884421 ----------UAAUGGAACCAGACCUCCGAUCCA-----GUCAGGAACCAAA-----UCGGAGGUCUGACUACACGUAACCCCUGCAAUUG-UCCAUGGCCUUCCCAUUUUUAAU-- ----------..(((((..((((((((((((((.-----....)))......-----))))))))))).......(((.....))).....-)))))((.....)).........-- ( -28.10, z-score = -3.07, R) >dp4.chr3 9125945 103 + 19779522 ----------GGAUUAAAGCAGACCUCCGACUCUGCUCUGUCGGGAACUGAGCUGAGUCGGAGGCCUGGCUGCAGCUAACACCAAGAGUUG-UCCAUGACCAUUCCA-UUUUAAU-- ----------(((....(((((.((((((((((.((((.((.....)).)))).)))))))))).)).)))((((((.........)))))-)..........))).-.......-- ( -39.40, z-score = -3.24, R) >droPer1.super_4 1179996 103 + 7162766 ----------GGAUUAAAGCAGACCUCCGACUCAGCUCUGUCGGGAACUGAACUGAGUCGGAGGCCUGGCUGCAGCUAACACCAAGAGUUG-UCCAUGACCAUUCCA-UUUUAAU-- ----------(((....(((((.((((((((((((.((.((.....)).)).)))))))))))).)).)))((((((.........)))))-)..........))).-.......-- ( -37.40, z-score = -3.14, R) >droWil1.scaffold_180701 1108610 90 + 3904529 ----------AUCUUGAGCUUGACCUCCGAUCCG-----AAUAGCAUUCGAA-----UCGGAGGUCUUGUGACAGUCAAGAUCAAGA-UAGGGCCAUGACAGGGACAUUUU------ ----------(((((((.(((((((((((((.((-----(((...))))).)-----))))))(((....))).)))))).))))))-).(..((......))..).....------ ( -35.90, z-score = -4.16, R) >droVir3.scaffold_12875 11293791 95 - 20611582 ------------AUGAAUGUAGACCUCCGAUUCAGG---GC--UUGACUGAA-----UCGGAGGUCUGGCCACAGUCGGCAUCAGCAGCUGUUCCUCUUCCACAGAUCCAUUUCCUU ------------..(((((.(((((((((((((((.---..--....)))))-----))))))))))((..(((((..((....)).))))).)).............))))).... ( -36.90, z-score = -3.24, R) >droMoj3.scaffold_6496 11237556 74 + 26866924 -------------UAAACGUAGACCUCCGAUUUGGC---GU-UUCAACCAAA-----UCGGAGGUCCGACCACAGUC------AGCA-CAGUUCCAUUUCCCU-------------- -------------.....((.((((((((((((((.---..-.....)))))-----))))))))).(((....)))------.)).-...............-------------- ( -27.70, z-score = -6.07, R) >droGri2.scaffold_15245 4890715 109 + 18325388 AGAGAUUCGCGGAUGAAUAUAGACCUCCGAUCCGGU---GUGGUAAUCCGAA-----UCGGAGGUCCAUCCACAAACGUAAGGAGCAGCUGUUCCAAGUCCACAUUCCCAUUCAAUC .(.((((.(.(((((......((((((((((.(((.---........))).)-----)))))))))))))).)........(((((....))))).)))))................ ( -36.20, z-score = -3.10, R) >consensus __________UGAUAAAAACAGACCUCCGACCCA_____GUCGGAAACCGAA_____UCGGAGGCCUGACAGCAGUUGACCCAACGAGUUG_UCCAUGGCCAUUCCA_UUUUAAU__ .......................(((((((.........((.....)).........)))))))..................................................... ( -9.85 = -9.84 + -0.01)

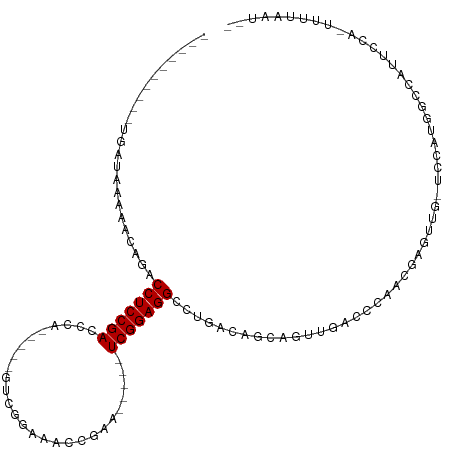
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:21 2011