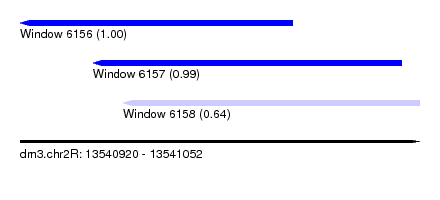
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,540,920 – 13,541,052 |
| Length | 132 |
| Max. P | 0.997276 |

| Location | 13,540,920 – 13,541,010 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.33 |
| Shannon entropy | 0.65241 |
| G+C content | 0.47676 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.29 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
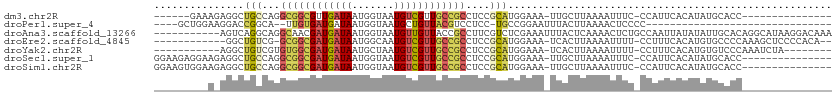
>dm3.chr2R 13540920 90 - 21146708 ------GAAAGAGGCUGCCAGGCGGCGUUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UUGCUUAAAAUUUC-CCAUUCACAUAUGCACC--------------- ------....(((((.((..((((((((((.......)))))))))))).))))).(((((((((-((......))))))-).........))))...--------------- ( -29.20, z-score = -1.96, R) >droPer1.super_4 1165268 75 + 7162766 ----GCUGGAAGGACCGGCA--UUGUGAUGAUAAUGGUAAUGCUGUUACGUCCUCC-UGCCGGAAUUUACUUAAAACUCCCC------------------------------- ----((.(((.(((((((((--(((..((....))..))))))))....)))))))-.)).(((.(((.....))).)))..------------------------------- ( -22.10, z-score = -2.66, R) >droAna3.scaffold_13266 9243281 102 + 19884421 -----------AGUCAGGCAGGCAACGAUGAUAAUGGUAAUGUUGUUACCGCCUUCGUCUCGAAAUUUACUCAAAACUCUGCCAAUUAUAUAUUGCACAGGCAUAAGGACAAA -----------.(((.((((((....(((((...(((((((...)))))))...)))))..((.......)).....))))))..........(((....)))....)))... ( -24.70, z-score = -1.55, R) >droEre2.scaffold_4845 7770174 96 - 22589142 ------------GGCUGUCG-GCGGCGAUGAUAAUGGCAAUGUCGUUGCCGCCUCCGCAUGGAAA-UCACUUAAAAUUUU-CCUUUCACAUGUGCCCCAAAGCUCCCCACA-- ------------((((...(-((((((((((((.......)))))))))))))..((((((((((-..............-..)))).))))))......)))).......-- ( -31.99, z-score = -2.66, R) >droYak2.chr2R 5496143 92 + 21139217 -----------AGGCUGUCGUGUGGCGAUGAUAAUGCUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UCACUUAAAAUUUU-CCUUUCACAUGUGUCCCAAAUCUA-------- -----------.((.....(.((((((((((((.......)))))))))))))..((((((((((-..............-..)))).))))))..)).......-------- ( -24.69, z-score = -2.45, R) >droSec1.super_1 11040377 96 - 14215200 GGAAGAGGAAGAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UUGCUUAAAAUUUC-CCAUUCACAUAUGCACC--------------- (((...((.((...)).)).(((((((((((((.......))))))))))))))))(((((((((-((......))))))-).........))))...--------------- ( -34.40, z-score = -2.51, R) >droSim1.chr2R 12277705 96 - 19596830 GGAAGUGGAAGAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UUGCUUAAAAUUUC-CCAUUCACAUAUGCACC--------------- ....(((((......(((.((((((((((((((.......))))))))))))))..))).(((((-((......))))))-)..))))).........--------------- ( -36.30, z-score = -2.96, R) >consensus _________A_AGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA_UUACUUAAAAUUUC_CCAUUCACAUAUGCACC_______________ ...............(((...((((((((((((.......))))))))))))....)))...................................................... (-13.09 = -13.29 + 0.19)
| Location | 13,540,944 – 13,541,046 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 62.11 |
| Shannon entropy | 0.74939 |
| G+C content | 0.49455 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.58 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.84 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13540944 102 - 21146708 UUAAUUAAGUGGCAAGCAGGAAGAGGAACA------GGAAGUGAAAGAGGCUGCCAGGCGGCGUUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UUGCUUA .....((((..(((.((.(((.........------((.(((.......))).)).(((((((.(((((.......))))).)))))))))))).))....-)..)))) ( -28.90, z-score = -0.45, R) >dp4.chr3 9111298 92 + 19779522 UUAAUUAAGUGCUCGGCUGGGCCUGGGCUG--------------GAAGGACCGGCAG--UGUGAUGAUAAUGGUAAUGCUGUUACGUCCUCCUCCCAGAAU-UUACUUA .....(((((((((....))))(((((..(--------------((.((((.(((((--(((.............))))))))..))))))).)))))...-..))))) ( -30.32, z-score = -1.48, R) >droAna3.scaffold_13266 9243321 94 + 19884421 UUAAUUAAGUGGCAUGCCG--AGCCGAACG-------------AGGCAGUCAGGCAGGCAACGAUGAUAAUGGUAAUGUUGUUACCGCCUUCGUCUCGAAAUUUACUCA .....(((((..(.((((.--.(((..((.-------------.....))..))).))))..(((((...(((((((...)))))))...)))))..)..))))).... ( -26.30, z-score = -1.03, R) >droEre2.scaffold_4845 7770211 87 - 22589142 --------GUAAUGAGUCGGGUGGCAGGCG------------GGAAGGGGCUGUC-GGCGGCGAUGAUAAUGGCAAUGUCGUUGCCGCCUCCGCAUGGAAA-UCACUUA --------....(((.(((.((((..((((------------(.......)))))-(((((((((((((.......))))))))))))).)))).)))...-))).... ( -34.70, z-score = -1.83, R) >droYak2.chr2R 5496174 94 + 21139217 UUAAUUAAGUGGCAGGCGGGCAGGCGAACA--------------GGGAGGCUGUCGUGUGGCGAUGAUAAUGCUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UCACUUA .....(((((((((.(((((...((((.((--------------(.....)))))))((((((((((((.......)))))))))))).))))).))....-))))))) ( -35.20, z-score = -1.86, R) >droSec1.super_1 11040401 108 - 14215200 UUAAUUAAGUGGCAGGCAGGAAGAGGAACAGGAAGUGGAAGAGGAAGAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UUGCUUA .....((((((((((.(...........((.....))...........).))))).(((((((((((((.......)))))))))))))(((....)))..-..))))) ( -34.55, z-score = -2.07, R) >droSim1.chr2R 12277729 108 - 19596830 UUAAUUAAGUGGCAGGCAGGAAGAGGAACAGGAAGUGGAAGUGGAAGAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA-UUGCUUA ........((((..(((((........((...........))........)))))((((((((((((((.......)))))))))))))))))).......-....... ( -34.79, z-score = -1.92, R) >droWil1.scaffold_180569 515508 94 - 1405142 CUAAUUAAGUGGUUGCCCAAGCUGAUGAUG--------------AUACAGUUGAU-GAUGAUUGUGAUGCGGAAAAUGUCGAAGUUGGUUUUUGGCUAGAAAUUACUUA .....((((((((((((.((((..((....--------------.(((((((...-...)))))))...(((......)))..))..))))..)))....))))))))) ( -19.00, z-score = -0.95, R) >consensus UUAAUUAAGUGGCAGGCAGGAAGAGGAACA______________AAGAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCCGCCUCCGCAUGGAAA_UUACUUA .....((((((((.(((................................))))))((((((((((((((.......))))))))))))))..............))))) (-15.79 = -15.58 + -0.21)
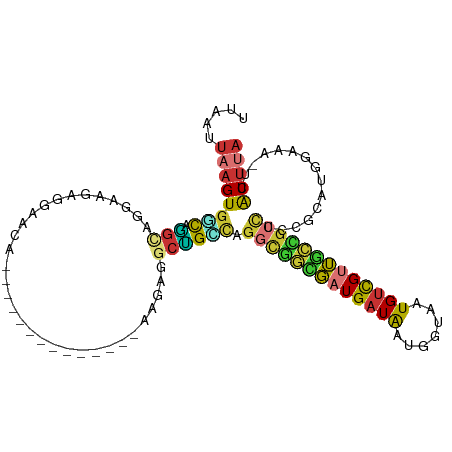
| Location | 13,540,954 – 13,541,052 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 63.68 |
| Shannon entropy | 0.67433 |
| G+C content | 0.52408 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -11.21 |
| Energy contribution | -10.46 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.90 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13540954 98 - 21146708 -----CAUCGGUUAAUUAAGUGGCAAGCAGGAAGAGGAACAGGAAGUGAAA------GAGGCUGCCAGGCGGCGUUGAUAAUGGUAAUGUCGUUGCC-GCCUCCGCAUGG -----((((((.........(((((.((..........((.....))....------...)))))))(((((((.(((((.......))))).))))-))).))).))). ( -30.53, z-score = -1.20, R) >dp4.chr3 9111308 88 + 19779522 -----CAUCAGUUAAUUAAGUGCUCGGCUGG-GCCUGGGCUGGA-------------AGGACCGGCAG--UGUGAUGAUAAUGGUAAUGCUGUUACG-UCCUCCUCCCAG -----..(((((..(........)..)))))-..(((((..(((-------------.((((.(((((--(((.............))))))))..)-)))))).))))) ( -27.92, z-score = -0.47, R) >droAna3.scaffold_13266 9243332 89 + 19884421 -----CAUCAGUUAAUUAAGUGGCAUGCCG--AGCCGAACGAGG-------------CAGUCAGGCAGGCAACGAUGAUAAUGGUAAUGUUGUUACC-GCCUUCGUCUCG -----..............((.((.(((((--((((......))-------------)..)).)))).)).))(((((...(((((((...))))))-)...)))))... ( -26.00, z-score = -1.08, R) >droYak2.chr2R 5496184 95 + 21139217 AGACGCGUCGGUUAAUUAAGUGGCAGGCGGGCAGGCGAACAGG--------------GAGGCUGUCGUGUGGCGAUGAUAAUGCUAAUGUCGUUGCC-GCCUCCGCAUGG ....((((((.((....)).)))).((.(((((.((((.(((.--------------....))))))).)((((((((((.......))))))))))-)))))))).... ( -35.20, z-score = -0.74, R) >droSec1.super_1 11040411 104 - 14215200 -----CAUCGUUUAAUUAAGUGGCAGGCAGGAAGAGGAACAGGAAGUGGAAGAGGAAGAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCC-GCCUCCGCAUGG -----..............((((..(((((...............................)))))((((((((((((((.......))))))))))-)))))))).... ( -33.98, z-score = -2.14, R) >droSim1.chr2R 12277739 104 - 19596830 -----CAUCGGUUAAUUAAGUGGCAGGCAGGAAGAGGAACAGGAAGUGGAAGUGGAAGAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCC-GCCUCCGCAUGG -----((((((.........((((((.(...........((.....))...........).))))))(((((((((((((.......))))))))))-))).))).))). ( -36.85, z-score = -2.82, R) >droWil1.scaffold_180569 515518 90 - 1405142 -----CUGCUGCUAAUUAAGUGGUUGCCCA--AGCUGAUGAUGA------------UACAGUUGAUGA-UGAUUGUGAUGCGGAAAAUGUCGAAGUUGGUUUUUGGCUAG -----..((..((.....))..)).(((.(--(((..((.....------------(((((((.....-.)))))))...(((......)))..))..))))..)))... ( -18.00, z-score = -0.16, R) >consensus _____CAUCGGUUAAUUAAGUGGCAGGCAGG_AGAGGAACAGGA_____________GAGGCUGCCAGGCGGCGAUGAUAAUGGUAAUGUCGUUGCC_GCCUCCGCAUGG .....................(((.(((................................))))))((((((((((((((.......)))))))))).))))........ (-11.21 = -10.46 + -0.74)

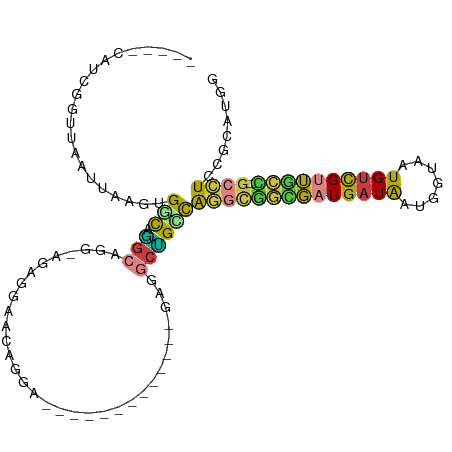
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:15 2011