| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,140,855 – 3,140,966 |
| Length | 111 |
| Max. P | 0.783086 |

| Location | 3,140,855 – 3,140,966 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.48 |
| Shannon entropy | 0.30395 |
| G+C content | 0.40509 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -22.61 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3140855 111 - 23011544 --------AUAUUUUAAAUCCAAGUGUAACCGCUGUCAAAAUGUCAAACGAGCUUAAAGUUGCCUUGCCAAAUUUUUGGGCGAAAAAGCCGCACAUGUGUGGCUUAGUCGAUAAAAGGA --------..........(((.((((....)))).......((((......(((((((.(((......)))...)))))))((..((((((((....))))))))..))))))...))) ( -28.00, z-score = -1.42, R) >droAna3.scaffold_12913 361144 103 - 441482 ------------CUUUUUUCUCAGUGUAGCACUUGUCAAAAUGUCAAACUGGCUUAAAUU----UUGGCGGGAUUUUGGGCGAAAGCUCCGCACAUGUGCGGCUUUUUCGAUAAAGGAC ------------(((((.((.......(((.((((((((((((((.....)))....)))----)))))))).....((((....))))((((....))))))).....)).))))).. ( -29.40, z-score = -1.75, R) >droEre2.scaffold_4929 3184642 118 - 26641161 ACCACUAAGUAUUUUAAAUCAAAGUGUAACCGCUGUCAAAAUGUCAAACGAGCUUAAAGUUGCCUUGCCAAA-AUUUGGGCGAAAAAGCCGCACAUGUGUGGCUUAGUCGAUAAAAGGA .((...................((((....)))).......((((......(((((((.(((......))).-.)))))))((..((((((((....))))))))..))))))...)). ( -27.20, z-score = -0.78, R) >droYak2.chr2L 3133590 118 - 22324452 ACUACUAAGUAUUUAAAAUCAAAGUGUAACCCCUGUCAAAAUGUCAAACGAGCUUAAAGUUGCCUUGCCAAA-UUUUGGGCGAAAAAGCCGCACAUGUGUGGCUUAGUCGAUAAAAGGU ............................(((..((((..............((((((((((.........))-))))))))((..((((((((....))))))))..))))))...))) ( -27.60, z-score = -1.44, R) >droSec1.super_5 1292578 110 - 5866729 --------AUAUUUAAAAUCCAAGUGUAACCGCUGUCAAAAUGUCAAACGAGCUUAAAGUUGCCUUGCCAAA-AUUUGGGCGAAAAAGCCGCACAUGUGUGGCUUAGUCGAUAAAAGGA --------..........(((.((((....)))).......((((......(((((((.(((......))).-.)))))))((..((((((((....))))))))..))))))...))) ( -27.80, z-score = -1.53, R) >droSim1.chr2L 3081979 110 - 22036055 --------AUAUUUAAAAUCCAAGUGUAACCGCUGUCAAAAUGUCAAACGAGCUUAAAGUUGCCUCGCCAAA-AUUUGGGCGAAAAAGCCGCACAUGUGUGGCUUAGUCGAUAAAAGGA --------..........(((.((((....)))).......((((....((((........).)))(((...-.....)))((..((((((((....))))))))..))))))...))) ( -28.60, z-score = -1.85, R) >consensus ________AUAUUUAAAAUCCAAGUGUAACCGCUGUCAAAAUGUCAAACGAGCUUAAAGUUGCCUUGCCAAA_AUUUGGGCGAAAAAGCCGCACAUGUGUGGCUUAGUCGAUAAAAGGA .............................((..((((..............(((((((.(((......)))...)))))))((..((((((((....))))))))..))))))...)). (-22.61 = -23.20 + 0.59)

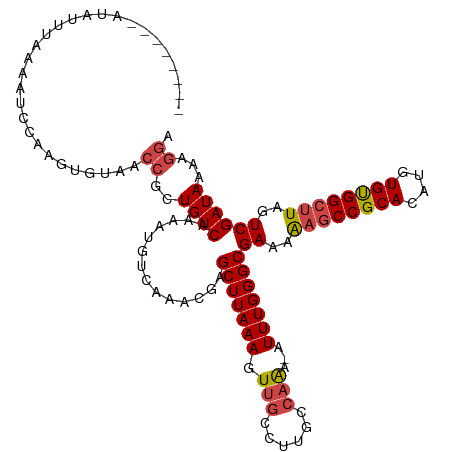

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:09 2011