
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,481,428 – 13,481,519 |
| Length | 91 |
| Max. P | 0.670214 |

| Location | 13,481,428 – 13,481,519 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 67.52 |
| Shannon entropy | 0.69484 |
| G+C content | 0.35186 |
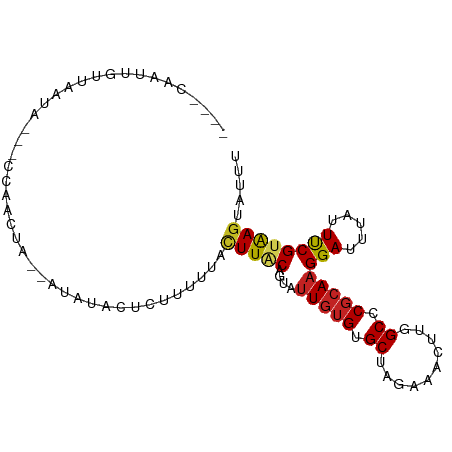
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -8.58 |
| Energy contribution | -8.53 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.38 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
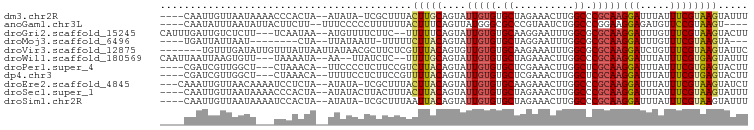
>dm3.chr2R 13481428 91 + 21146708 ----CAAUUGUUAAUAAAACCCACUA--AUAUA-UCGCUUUACUUGCAGUAUUGUGUGCUAGAAACUUGGCCCGCAAGGAUUUAUUUCGUAAGUAUUU ----......................--.....-......(((((((....(((((.(((((....))))).)))))(((.....))))))))))... ( -16.90, z-score = -0.79, R) >anoGam1.chr3L 22176284 88 - 41284009 ----CAAUAUUUAAUAUUACUUCUU--UUUCCCCCUUUUUUACUUUCAGUUAUGGGCGCCCGUAAUCUGGCCCGGAAGGAGAUGUUCCGUAAGU---- ----....(((((.....((.((((--(((((.............((((((((((....)))))).))))...))))))))).))....)))))---- ( -18.99, z-score = -0.92, R) >droGri2.scaffold_15245 4787692 91 - 18325388 CAUUUGAUUGUCUCUU---UCAAUAA--AUGUUUUCUUC--UUUUUCAGUAUUGUGUGCAAGGAAUUUGGCGCGCAAGGAUUUGUUUCGUAAGUACUU ((((((.(((......---.))))))--)))........--......((((((((((((..........))))))..(((.....)))...)))))). ( -14.20, z-score = 0.30, R) >droMoj3.scaffold_6496 11134089 80 - 26866924 ----UGAUUAUUAAU--------CUA--UUAUAAUU-UUUUUCUUACAGUAUUGUGUGCUAGGAAUUUGGCGCGCAAGGAUUUGUUUCGUAAGUA--- ----...........--------...--........-.....(((((....(((((((((((....)))))))))))(((.....))))))))..--- ( -17.70, z-score = -2.06, R) >droVir3.scaffold_12875 11199408 91 + 20611582 -------UGUUUGAUAUUGUUUAUUAAUUAUAACGCUUCUCGUUUACAGUGUUGUGUGCAAGAAAUUUGGCGCGCAAGGAUCUGUUUCGUAAGUAUUC -------...((((((.....)))))).......((((..((...(((((.((((((((.(....)...)))))))).)..))))..)).)))).... ( -19.00, z-score = -0.74, R) >droWil1.scaffold_180569 437103 89 + 1405142 CAAUUAAUUAAGUGUU---UAAAAUA--AA--UUAUCUC--UUUUGCAGUAUUGUGUGCUAGAAACUUGGCCCGCAAGGAUUUAUUUCGUGAGUAUUU .........(((((((---(((((((--((--((.....--..........(((((.(((((....))))).))))).)))))))))..))))))))) ( -12.99, z-score = 0.29, R) >droPer1.super_4 1108568 89 - 7162766 ----CGAUCGUUGGCU---CUAAACA--UUCCCCUCUUCCGUCUUACAGUAUUGUGUGCUCGAAACUUGGCUCGCAAGGAUUUAUUUCGUGAGUACUU ----.((..(((....---...))).--.)).........(((((((....(((((.(((........))).)))))(((.....)))))))).)).. ( -15.00, z-score = 0.74, R) >dp4.chr3 9048606 89 - 19779522 ----CGAUCGUUGGCU---CUAAACA--UUUUCCUCUUCCGUUUUACAGUAUUGUGUGCUCGAAACUUGGCUCGCAAGGAUUUAUUUCGUGAGUACUU ----.........(((---(......--...(((......(.....)....(((((.(((........))).))))))))..........)))).... ( -13.11, z-score = 1.29, R) >droEre2.scaffold_4845 7709936 92 + 22589142 ---CAAAUUGUUAACAAAAUCCUCUA--AUAUA-UCGCUUUACUUACAGUAUUGUGUGCAAGAAACUUGGCCCGCAAGGAUUUAUUUCGUAAGUAUCU ---.......................--.....-......(((((((.((((...))))..((((...(..((....))..)..)))))))))))... ( -13.90, z-score = -0.33, R) >droSec1.super_1 10980737 92 + 14215200 ----CAAUUGUUAAUAAAACCCACUA--AUAUACUUACUUUACUUACAGUAUUGUGUGCUAGAAACUUGGCCCGCAAGGAUUUAUUUCGUAAGUAUUU ----......................--..((((((((.............(((((.(((((....))))).)))))(((.....))))))))))).. ( -17.00, z-score = -1.31, R) >droSim1.chr2R 12217338 91 + 19596830 ----CAAUUGUUAAUAAAAUCCACUA--AUAUA-UCGCUUUAAUUACAGUAUUGUGUGCUAGAAACUUGGCCCGCAAGGAUUUAUUUCGUAAGUAUUU ----............((((((....--.....-..(((........))).(((((.(((((....))))).)))))))))))............... ( -15.00, z-score = -0.61, R) >consensus ____CAAUUGUUAAUA___CCAACUA__AUAUACUCUUUUUACUUACAGUAUUGUGUGCUAGAAACUUGGCCCGCAAGGAUUUAUUUCGUAAGUAUUU ..........................................(((((....(((((.((..........)).)))))(((.....))))))))..... ( -8.58 = -8.53 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:07 2011