| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,451,405 – 13,451,506 |
| Length | 101 |
| Max. P | 0.843514 |

| Location | 13,451,405 – 13,451,506 |
|---|---|
| Length | 101 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.95 |
| Shannon entropy | 0.66460 |
| G+C content | 0.47803 |
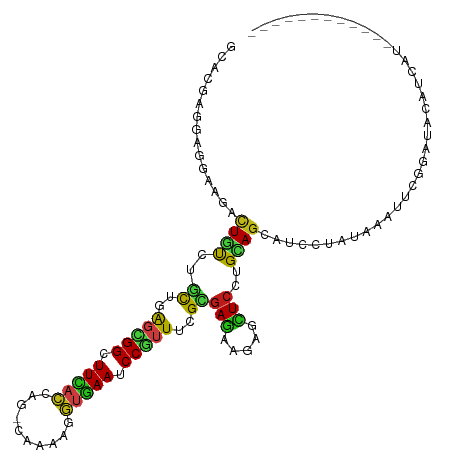
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -11.90 |
| Energy contribution | -10.99 |
| Covariance contribution | -0.90 |
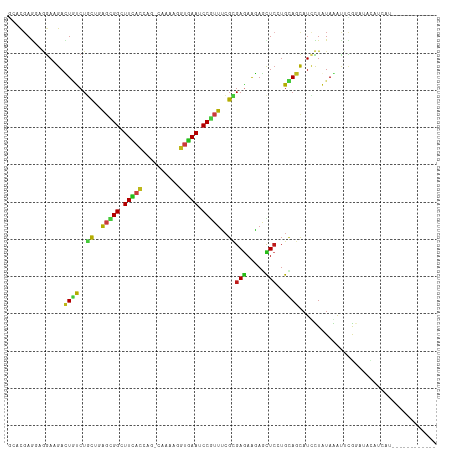
| Combinations/Pair | 1.79 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
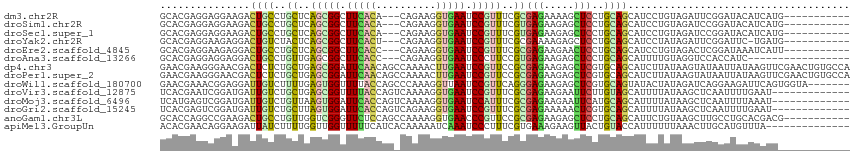
>dm3.chr2R 13451405 101 + 21146708 GCACGAGGAGGAAGACUGCCUGCUCAGCGGCUUCACA---CAGAAGGUGAAUCCGUUUCGCGAGAAAAGCUCCUGCAGCAUCCUGUAGAUUCGGAUACAUCAUG----------- ((..(((.(((.......))).))).)).........---.....((((.(((((((((....)))).....((((((....))))))...))))).))))...----------- ( -30.40, z-score = -0.44, R) >droSim1.chr2R 12187479 101 + 19596830 GCACGAGGAGGAAGACUGCCUGCUCAGCGGCUUCACA---CAGAAGGUGAAUCCGUUUCGUGAGAAGAGCUCCUGCAGCAUCCUGUAGAUCCGGAUACAUCAUG----------- ....((..((((.(.((((..((((.((((.(((((.---......))))).))))(((....)))))))....))))).))))(((........))).))...----------- ( -32.00, z-score = -0.53, R) >droSec1.super_1 10950853 101 + 14215200 GCACGAGGAGGAAGACUGCCUGCUCAGCGGCUUCACA---CAGAAGGUGAAUCCGUUUCGUGAGAAGAGCUCCUGCAGCAUCCUGUAGAUCCGGAUACAUCAUG----------- ....((..((((.(.((((..((((.((((.(((((.---......))))).))))(((....)))))))....))))).))))(((........))).))...----------- ( -32.00, z-score = -0.53, R) >droYak2.chr2R 5403693 100 - 21139217 GCACGAGGAAGAGGACUGUCUACUCAGCGGCUUCACU---CAGAAGGUGAAUCCGUUUCGCGAAAAGAGCUCCUGCAGCAUCCUAUAGAUUCGGAUUC-UGAUG----------- ....(((((((..(.(((......))))..)))).))---).......(((((((..((.......(((((.....))).)).....))..)))))))-.....----------- ( -25.70, z-score = 0.86, R) >droEre2.scaffold_4845 7678698 101 + 22589142 GCACGAGGAAGAGGACUGCCUGCUCAGCGGCUUCACC---CAGAAGGUGAAUCCGUUUCGCGAGAAGAACUCCUGCAGCAUCCUGUAGACUCGGAUAAAUCAUU----------- ...((((...(((..((.(.(((..(((((.((((((---.....)))))).)))))..))).).))..)))((((((....)))))).))))...........----------- ( -34.80, z-score = -1.81, R) >droAna3.scaffold_13266 9086872 95 - 19884421 GCACGAGGAGGAGGACUGCCUGUUGAGCGGCUUCACC---CAGAAGGUGAAUCCCUUCCGUGAGAAGAGCUCCUGCAGCAUUUUGUAGGUCCACCAUC----------------- .........((.(((((((((((.((((((.((((((---.....)))))).))(((.(....)))).))))..))))......))).)))).))...----------------- ( -32.10, z-score = -0.80, R) >dp4.chr3 5351257 115 + 19779522 GAACGAAGGGAACGACUCUCUGCUGAGCGGAUUCAACAGCCAAAACUUGAAUCCGUUCCGCGAGAAGAGCUCGUGCAGCAUCUUAUAAGUAUAAUUAUAAGUUCGAACUGUGCCA .......((..((((((((((((.((((((((((((..........)))))))))))).)).)).)))).))))((((...(((((((......)))))))......)))).)). ( -38.50, z-score = -3.47, R) >droPer1.super_2 5556496 115 + 9036312 GAACGAAGGGAACGACUCUCUGCUGAGCGGAUUCAACAGCCAAAACUUGAAUCCGUUCCGCGAGAAGAGCUCGUGCAGCAUCUUAUAAGUAUAAUUAUAAGUUCGAACUGUGCCA .......((..((((((((((((.((((((((((((..........)))))))))))).)).)).)))).))))((((...(((((((......)))))))......)))).)). ( -38.50, z-score = -3.47, R) >droWil1.scaffold_180700 4788141 108 + 6630534 GAACGAAACGGAGGAUUGUCUUUUGAGUGGUUUUACCAGCCCAAAGGUUAAUCCGUUCAGGGAGAAGAGCUCGUGCAGUAUACUAUAGAUCAGGAAGAUUCAGUGGUA------- ..((...((((.((((((.((((((.((((.....))).).)))))).))))))((((........))))))))...)).((((((.((((.....))))..))))))------- ( -27.80, z-score = -0.91, R) >droVir3.scaffold_12875 4543756 102 + 20611582 UCACGAAUCGGAUGAUUGUCUGCUGAGCGGUUUUACCAGUCAAAAGGUGAAUCCGUUUCGCGAGAAGAAUUCUUGUAGCAUUUUAUAAGCUCAAUUUUGAAU------------- ...((((..((((..((.((.((.((((((.((((((........)))))).)))))).)))).))..))))(((.(((.........)))))).))))...------------- ( -26.50, z-score = -1.35, R) >droMoj3.scaffold_6496 1339035 102 - 26866924 UCAUGAGUCGGAUGAUUGUCUGUUAAGUGGAUUCACCAGUCAAAAGGUGAAUCCAUUUCGCGAGAAGAAUUCAUGCAGCAUUUUAUAAGCUCAAUUUUAAAU------------- ...(((((...((((.((.((((...(((((((((((........)))))))))))(((....)))........))))))..))))..))))).........------------- ( -31.30, z-score = -3.92, R) >droGri2.scaffold_15245 17910594 102 - 18325388 UCACGAGUCGGAUGAUUGUCUGCUUAGUGGAUUCACCAGUCAGAAGGUGAAUCCGUUUCGCGAGAAAAACUCGUGCAGCAUUUUAUAAGCUCAAUUUUGAAU------------- .......(((((..((((...(((((..(((((((((........)))))))))(((.((((((.....)))))).)))......))))).)))))))))..------------- ( -34.40, z-score = -3.43, R) >anoGam1.chr3L 5195326 104 + 41284009 GCACCAGGCCGAAGACUGCCUGUUGGUCGGGUUCUCCAGCCAAAAGGUGAACCCGUUCCGCGAGAAGAGCUCCUGCAGCAUUCUGUAAGCUUGCCUGCACGACG----------- .((.(((((........))))).))(((((((((.((........)).)))))).....((.((..(((((..(((((....))))))))))..))))..))).----------- ( -38.60, z-score = -1.62, R) >apiMel3.GroupUn 246351871 101 - 399230636 ACACGAACAGGAAGAUUAUCUUUUGGUUGGUUUUUCAUCACAAAAAUCAAAUCCCUUUCGUGAAAGAAGUUACUGUACCAUUUUUUAAACUUGCAUGUUUA-------------- .((((((.((((((.....)))....(((((((((......)))))))))...))))))))).......................(((((......)))))-------------- ( -16.70, z-score = -0.16, R) >consensus GCACGAGGAGGAAGACUGUCUGCUGAGCGGCUUCACCAG_CAAAAGGUGAAUCCGUUUCGCGAGAAGAGCUCCUGCAGCAUCCUAUAAAUUCGGAUACAUCAU____________ ...............((((..((..(((((.(((((..........))))).)))))..))(((.....)))..))))..................................... (-11.90 = -10.99 + -0.90)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:04 2011