| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,426,348 – 13,426,445 |
| Length | 97 |
| Max. P | 0.512489 |

| Location | 13,426,348 – 13,426,445 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 60.20 |
| Shannon entropy | 0.80027 |
| G+C content | 0.53792 |
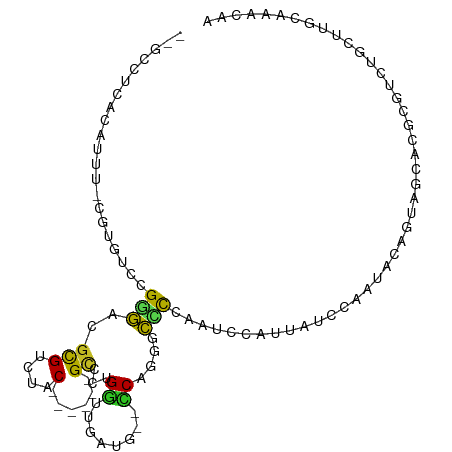
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -1.77 |
| Energy contribution | -1.39 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 2.31 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.05 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.512489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
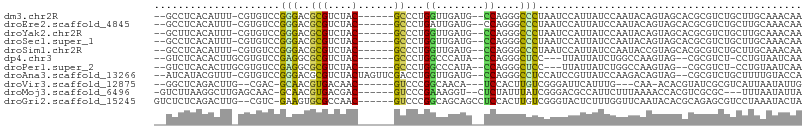
>dm3.chr2R 13426348 97 + 21146708 --GCCUCACAUUU-CGUGUCCGGGACGCGUCUAC------GCCCUGGUUGAUG--CCAGGGCCCUAAUCCAUUAUCCAAUACAGUAGCACGCGUCUGCUUGCAAACAA --((..(((....-.)))....((((((((((((------((((((((....)--)))))))........(((....)))...)))).))))))))....))...... ( -34.90, z-score = -3.29, R) >droEre2.scaffold_4845 7653573 97 + 22589142 --GCCUCACAUUU-CGUGUCCGGGACGCGUCUAC------GCCCUGAUUGAUG--CCAGGGCCCUAAUCCAUUAUCCAAUACAGUAGCACGCGUCUGCUUGCAAACAA --((..(((....-.)))....((((((((((((------((((((.......--.))))))........(((....)))...)))).))))))))....))...... ( -29.80, z-score = -2.15, R) >droYak2.chr2R 5378368 97 - 21139217 --GCUUCACAUUU-CGUGUCCGGGACGCGUCUAC------GCCCUGGUUGAUG--CCAGGGCCCUAAUCCAUUAUCCAAUACAGUAGCACGCGUCUGCUUGCAAACAA --((..(((....-.)))....((((((((((((------((((((((....)--)))))))........(((....)))...)))).))))))))....))...... ( -35.30, z-score = -3.47, R) >droSec1.super_1 10926130 97 + 14215200 --GCCUCACAUUU-CGUGUCCGGGACGCGUCUAC------GCCCUGGUUGAUG--CCAGGGCCCUAAUCCAUUAUCCAAUACAGUAGCACGCGUCUGCUUGCAAACAA --((..(((....-.)))....((((((((((((------((((((((....)--)))))))........(((....)))...)))).))))))))....))...... ( -34.90, z-score = -3.29, R) >droSim1.chr2R 12163357 97 + 19596830 --GCCUCACAUUU-CGUGUCCGGGACGCGUCUAC------GCCCUGGUUGAUG--CCAGGGCCCUAAUCCAUUAUCCAAUACCGUAGCACGCGUCUGCUUGCAAACAA --((..(((....-.)))....((((((((((((------((((((((....)--)))))))........(((....)))...)))).))))))))....))...... ( -35.00, z-score = -3.31, R) >dp4.chr3 5323489 92 + 19779522 --GUCUCACACUUGCGUGUCCGAGGCGCGUCUAC------GCCCUGGCCCAUA--CCAGGGCUCC---UUAUUAUCUGGCCAAGUAG--CGCGUCU-CCUGUAAUCAA --.....((((....))))..((((((((.((((------(((((((......--))))))).((---.........))....))))--)))))))-).......... ( -36.60, z-score = -4.21, R) >droPer1.super_2 5528686 92 + 9036312 --GUCUCACACUUGCGUGUCCGAGGCGCGUCUAC------GCCCUGGCCCAUA--CCAGGGCUCC---UUAUUAUCUGGCCAAGUAG--CGCGUCU-CCUGUAAUCAA --.....((((....))))..((((((((.((((------(((((((......--))))))).((---.........))....))))--)))))))-).......... ( -36.60, z-score = -4.21, R) >droAna3.scaffold_13266 5136443 101 - 19884421 --AUCAUACGUUU-CGUGUCCGGGACGCGUCUACUAGUUCGACCUGGUUGAUG--CCAGGGCCUCCAUCCGUUAUCCAAGACAGUAG--CGCGUCUGCUUUUGUACCA --...(((((...-)))))..((((((((.(((((.......((((((....)--)))))..........(((......))))))))--))))))(((....))))). ( -31.80, z-score = -1.51, R) >droVir3.scaffold_12875 10087957 90 - 20611582 --GGCUCAGACUUG--CGAC-GCAACGUGACAAC------GUCCCGGCAACA---UCCACUUGUCGGGAUUCAUUUG---CAA-ACACGUAUCGCGUCAUUAAUAUUG --.((........)--)(((-((.(((((.(((.------((((((((((..---.....))))))))))....)))---...-.)))))...))))).......... ( -28.50, z-score = -2.93, R) >droMoj3.scaffold_6496 16453507 95 + 26866924 -GUCUUAAGGCUUGAGCAAC-GCAACGUGACGAC------GUCCCGAAAGGU--CUCUAUUUAUCGGGACGCCAUUCUUUAAAACCACGUCGCGC---UUUAAUAUUA -...(((((((....((...-))...((((((.(------(((((((.((((--....)))).))))))))...((.....))....))))))))---)))))..... ( -26.40, z-score = -1.98, R) >droGri2.scaffold_15245 14681141 99 - 18325388 GUCUCUCAGACUUG--CGUC-GAAGUGCGCCAAC------GUCCCGGCAGCAGCCUCCACUUGUCGGGUACUCUUUGGUUCAAUACACGCAGAGCGUCCUAAAUACUA (((.....)))(((--(((.-....((.(((((.------((((((((((..........)))))))).))...))))).))....))))))................ ( -26.40, z-score = -0.40, R) >consensus __GCCUCACAUUU_CGUGUCCGGGACGCGUCUAC______GCCCUGGUUGAUG__CCAGGGCCCCAAUCCAUUAUCCAAUACAGUAGCACGCGUCUGCUUGCAAACAA ...............((((.....))))................(((.(((((.....(....).....))))).))).............................. ( -1.77 = -1.39 + -0.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:02 2011