
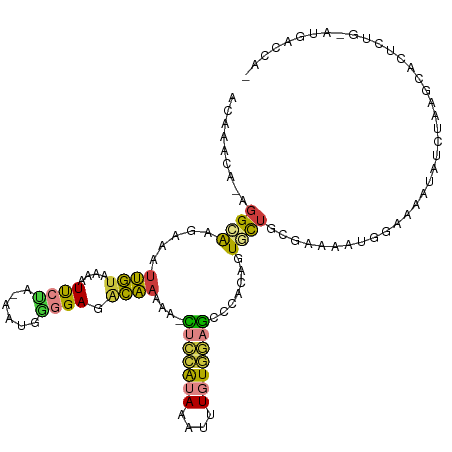
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,139,947 – 3,140,080 |
| Length | 133 |
| Max. P | 0.796368 |

| Location | 3,139,947 – 3,140,051 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.16 |
| Shannon entropy | 0.71026 |
| G+C content | 0.35650 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -7.31 |
| Energy contribution | -6.79 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3139947 104 + 23011544 ACAAACA-AGGCAAGAAAUUGUAAAAUUCUA-AAUGAGGAGACAAAAAACUCCAUAAAUUUGUGGAGUCCACAGUGCUGAAAAAGUGGAAAACAUGGCAUGACCAU---------- .......-..((((....)))).........-.....(....)......(((((((....)))))))(((((..(......)..)))))....((((.....))))---------- ( -20.90, z-score = -1.42, R) >droSim1.chr2L 3081074 113 + 22036055 ACAAACA-AGGCAAGAAAUUGUAAAAUUCUA-AAUGGGGAGACAAAAA-CUCCAUAAAUUUGUGGAGCUCACAGUGCUGCGAAAGUGGAAAACAUCUAAGCACUCUGAAUGACCAU .......-..((((....)))).........-.(((((....).....-(((((((....))))))).(((.((((((((....))(((.....))).)))))).)))....)))) ( -29.20, z-score = -2.96, R) >droSec1.super_5 1291682 113 + 5866729 ACAAACA-AGGCAAGAAAUUGUAAAAUUCUA-AAUGGGGAGACAAAAA-CUCCAUAAAUUUGUGGAGCCCGUAGUGCUGCGAAAGUGGAAAACAUUUAAGCACUCUGAAUGACCAU .......-..((((....))))...((((..-.(((((....).....-(((((((....))))))).))))((((((....(((((.....))))).))))))..))))...... ( -28.00, z-score = -2.32, R) >droYak2.chr2L 3132700 113 + 22324452 ACAAACA-AGGCAAGAAAUUGUAAAAUUCUA-AAUGGUGAGGCAAAAA-CUCCAUAAAUUUGUGGAGUCCACAGUGCUGCGAAAAUGGAAAUUAUCUAAGCACUCUGCAUGACCAA .......-..((((....)))).........-..((((.(.(((...(-(((((((....))))))))....((((((..((.(((....))).))..)))))).))).).)))). ( -31.30, z-score = -3.27, R) >droEre2.scaffold_4929 3183779 113 + 26641161 ACAAACA-AGGAAAGAAAUUGUAAAAUUCUA-AAUGGGGAGGCAAAAA-CUCCAUAAAUUUGUGGAGUCCACAGUGCUGCGAAAAUGGAAAUUAUCUCAGCACUCUGCAUGACCAA ....(((-(.........)))).........-..(((....(((...(-(((((((....))))))))....(((((((.((.(((....))).)).))))))).)))....))). ( -30.60, z-score = -2.92, R) >droAna3.scaffold_12913 360318 93 + 441482 ACAAGCACAGGAAAGAAAUUGUAAAAUCCAACGAAUGGGAAGCAAAAAGGCUAGCAAAUUUAUGAACCCUACGGUUUUACAGAAAUUGUUUUU----------------------- ..................((((((((((........(((.(((......)))..((......))..)))...))))))))))...........----------------------- ( -11.90, z-score = 0.81, R) >droVir3.scaffold_12963 20090601 85 + 20206255 ------UCGAGCGGGAUCGAAUGUAAUUCAUUAAAUUUGAAUUUGAAGAGCUAAGAAAAUUAUUUUUCUCAGAAUAU-----AAACGGAUAAUCUA-------------------- ------((((((....((((((..((((.....))))...))))))...))).((((((....))))))........-----...)))........-------------------- ( -10.30, z-score = 1.09, R) >consensus ACAAACA_AGGCAAGAAAUUGUAAAAUUCUA_AAUGGGGAGACAAAAA_CUCCAUAAAUUUGUGGAGCCCACAGUGCUGCGAAAAUGGAAAAUAUCUAAGCACUCUG_AUGACCA_ .........((((.....((((....((((......)))).))))....(((((((....))))))).......))))...................................... ( -7.31 = -6.79 + -0.52)


| Location | 3,139,983 – 3,140,080 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 59.27 |
| Shannon entropy | 0.80385 |
| G+C content | 0.36195 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -4.26 |
| Energy contribution | -4.16 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3139983 97 + 23011544 GAGACAAAAAACUCCAUAAAUUUGUGGAGUCCACAGUGCUGAAAAAGUGGAAAACAUG----------GCAUGACC-AUAAGUUGCCCUAUAAAAUGCUCAAAAUAAA (((.((.....(((((((....)))))))(((((..(......)..)))))......(----------(((.....-......))))........)))))........ ( -21.40, z-score = -2.40, R) >droSim1.chr2L 3081110 106 + 22036055 GAGACAAAAA-CUCCAUAAAUUUGUGGAGCUCACAGUGCUGCGAAAGUGGAAAACAUCUAAGCACUCUGAAUGACC-AUAAGUUGCCCUAUAAAAUGCUCUAAAUAAA (((.((....-.......(((((((((...(((.((((((((....))(((.....))).)))))).)))....))-)))))))...........)))))........ ( -24.65, z-score = -2.80, R) >droSec1.super_5 1291718 106 + 5866729 GAGACAAAAA-CUCCAUAAAUUUGUGGAGCCCGUAGUGCUGCGAAAGUGGAAAACAUUUAAGCACUCUGAAUGACC-AUAAGUUGCCCUAUAAAAUGCUUAAAAUUAA (((.((....-(((((((....)))))))...((((.((.((....((((....((((((.......)))))).))-))..)).)).))))....)))))........ ( -20.70, z-score = -1.64, R) >droYak2.chr2L 3132736 106 + 22324452 GAGGCAAAAA-CUCCAUAAAUUUGUGGAGUCCACAGUGCUGCGAAAAUGGAAAUUAUCUAAGCACUCUGCAUGACC-AAGAAUUGCCCUAUAAAAUUCUCAAAAUAAA ..(((((..(-(((((((....)))))))).((.((((((..((.(((....))).))..)))))).)).......-.....)))))..................... ( -26.80, z-score = -3.42, R) >droEre2.scaffold_4929 3183815 106 + 26641161 GAGGCAAAAA-CUCCAUAAAUUUGUGGAGUCCACAGUGCUGCGAAAAUGGAAAUUAUCUCAGCACUCUGCAUGACC-AACAAUUGCCCUGUAAAAUGCUCAAAAUAAA ..(((((..(-(((((((....)))))))).((.(((((((.((.(((....))).)).))))))).)).......-.....)))))..................... ( -30.90, z-score = -4.05, R) >droAna3.scaffold_12913 360357 85 + 441482 -AAGCAAAAAGGCUAGCAAAUUUAUGAACCCUACGGUUUUACAGAAAUUGUUUUUAAGU------------UGUCCGAUCUAUUCGACUUCAAUACAC---------- -.(((......)))(((((.(((..(((((....)))))....))).))))).....((------------((..(((.....)))....))))....---------- ( -10.60, z-score = 0.41, R) >droVir3.scaffold_12963 20090634 90 + 20206255 -AAUUUGAAGAGCUAAGAAAAUUAUUUUUCUCAGAAUA-UAAACGGAUAAUCUAAAGCAGUGCCAUAGGGUAAUCUGAAUCAAGGCAUUGCA---------------- -.(((((.....((.((((((....)))))).))....-....)))))........((((((((....(((.......)))..)))))))).---------------- ( -18.52, z-score = -1.39, R) >consensus GAGACAAAAA_CUCCAUAAAUUUGUGGAGCCCACAGUGCUGCGAAAAUGGAAAACAUCUAAGCACUCUGCAUGACC_AUAAAUUGCCCUAUAAAAUGCUCAAAAUAAA ...........(((((((....)))))))............................................................................... ( -4.26 = -4.16 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:08 2011