
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,307,961 – 13,308,052 |
| Length | 91 |
| Max. P | 0.695505 |

| Location | 13,307,961 – 13,308,052 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.04 |
| Shannon entropy | 0.69859 |
| G+C content | 0.38578 |
| Mean single sequence MFE | -19.99 |
| Consensus MFE | -11.00 |
| Energy contribution | -10.07 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.35 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695505 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
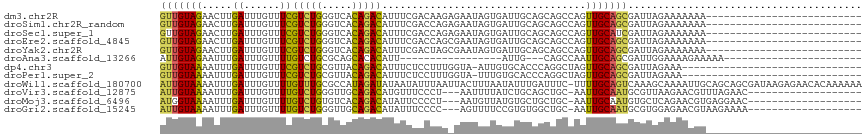
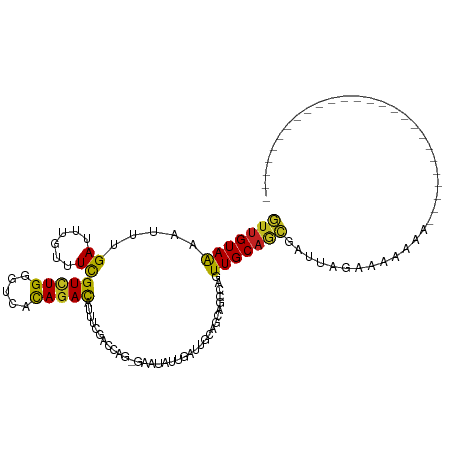
>dm3.chr2R 13307961 91 - 21146708 GUUGUAGAACUUGAUUUGUUUCGUCUGGGUCACAGACAUUUCGACAAGAGAAUAGUGAUUGCAGCAGCCAGUUGCAGCGAUUAGAAAAAAA-------------------------- .((((.(((...((......))(((((.....)))))..))).))))........(((((((.((((....)))).)))))))........-------------------------- ( -21.80, z-score = -0.78, R) >droSim1.chr2R_random 70620 91 - 2996586 GUUGUAGAACUUGAUUUGUUUCGUCUGGGUCACAGACAUUUCGACCAGAGAAUAGUGAUUGCAGCAGCCAGUUGCAGCGAUUAGAAAAAAA-------------------------- ..................(((((((((.....))))).((((.....))))....(((((((.((((....)))).)))))))))))....-------------------------- ( -21.60, z-score = -0.71, R) >droSec1.super_1 10809114 91 - 14215200 GUUGUAGAACUUGAUUUGUUUCGUCUGGGUCACAGACAUUUCGACCAGAGAAUAGUGAUUGCAGCAGCCAGUUGCAUCGAUUAGAAAAAAA-------------------------- ..................(((((((((.....))))).((((.....)))).(((((((.(((((.....)))))))).))))))))....-------------------------- ( -19.20, z-score = -0.03, R) >droEre2.scaffold_4845 10046725 91 + 22589142 GUUGUAGAACUUGAUUUGUUUCGUCUGGGUCACAGACAUUUCGACCAGCGAAUAGUGAUUGCAGCAGCCAGUUGCAGCGAUUAGAAAAAAA-------------------------- ((((..(((...((......))(((((.....)))))..)))...))))......(((((((.((((....)))).)))))))........-------------------------- ( -22.90, z-score = -0.83, R) >droYak2.chr2R 2978739 91 + 21139217 GUUGUAGAACUUGAUUUGUUUCGUCUGGGUCACAGACAUUUCGACUAGCGAAUAGUGAUUGCAGCAGCCAGUUGCAGCGAUUAGAAAAAAA-------------------------- ..................(((((((((.....)))))..((((.....))))...(((((((.((((....)))).)))))))))))....-------------------------- ( -21.80, z-score = -0.37, R) >droAna3.scaffold_13266 8803150 74 - 19884421 AUUGUAGAAUUUGAUUUGUUUUGUCUGCGCAGCACACAUU-----------------AUUG---CAGCCAAUUGCAGCGAUUGGAAAAGAAAAA----------------------- .((((((....((((.(((.((((....)))).))).)))-----------------))))---)))(((((((...)))))))..........----------------------- ( -12.20, z-score = 1.37, R) >dp4.chr3 4771289 86 - 19779522 GUUGUAAAAUUUGAUUUGUUUCGUCUGCGUUACAGACAUUUCUCCUUUGGUA-AUUGUGCACCCAGGCUAGUUGCAGCGAUUAGAAA------------------------------ (((((((...............(((((.....)))))......(((..(((.-.......))).)))....))))))).........------------------------------ ( -16.60, z-score = 0.15, R) >droPer1.super_2 4965200 86 - 9036312 GUUGUAAAAUUUGAUUUGUUUCGUCUGCGUUACAGACAUUUCUCCUUUGGUA-UUUGUGCACCCAGGCUAGUUGCAGCGAUUAGAAA------------------------------ (((((((.....((......))(((((.((((((((.(((........))).-)))))).)).)))))...))))))).........------------------------------ ( -18.50, z-score = -0.57, R) >droWil1.scaffold_180700 4552264 116 - 6630534 AUUGUAAAAUUUGAUUUGUUUUGUUUGCGCCAUAGAUAUAAUAUUUAAUUACUUUAAUAUUUGAUUUC-UUUUGCAGUCAAAGCAAAAUUGCAGCAGCGAUAAGAGAACACAAAAAA .((((.........(((((((((.(((((....(((...((((((..........)))))).....))-)..))))).)))))))))(((((....)))))........)))).... ( -20.30, z-score = -0.10, R) >droVir3.scaffold_12875 7746646 94 + 20611582 AUUGUAAAAUUUGAUUUGUUUUGUCUGGGUUGCAGACAUGUUUCCCU---AAUUUUAUCUGCAGCUGC-AAUUGCAAUGCGUUAAGAACGUUUAGAAC------------------- .........(((((..((((((....(((..(((....)))..))).---..........(((..(((-....))).)))....)))))).)))))..------------------- ( -18.80, z-score = -0.27, R) >droMoj3.scaffold_6496 6740107 94 + 26866924 AUGGUAAAAUUUGAUUUGUUUUGUCUGUGUCACAGACAUAUUCCCCU---AAUGUUAUGUGCUGCUGC-AAUUGCAAUGUGCUCAGAACGUGAGGAAC------------------- .....................((((((.....))))))......(((---.(((((.((.((...(((-....)))....)).)).))))).)))...------------------- ( -21.60, z-score = -0.59, R) >droGri2.scaffold_15245 7504989 94 - 18325388 AUUGUAAAAUUUGAUUUGUUUUGUCUGGGUUGCAGACAUAUUUCCCC---AGUUUUCCGUGUGGCUGC-AAUUGCAAUGCGUGGAGAACGUAAGAAAA------------------- .....................((((((.....))))))..((((...---.((((((((((((..(((-....))).))))))))))))....)))).------------------- ( -24.60, z-score = -1.41, R) >consensus GUUGUAAAAUUUGAUUUGUUUCGUCUGGGUCACAGACAUUUCGACCAG_GAAUAUUGAUUGCAGCAGCCAGUUGCAGCGAUUAGAAAAAAA__________________________ (((((((.....((......))(((((.....)))))..................................)))))))....................................... (-11.00 = -10.07 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:48 2011