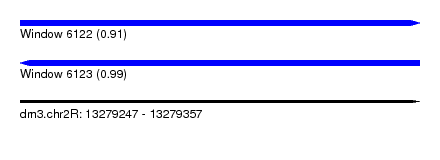
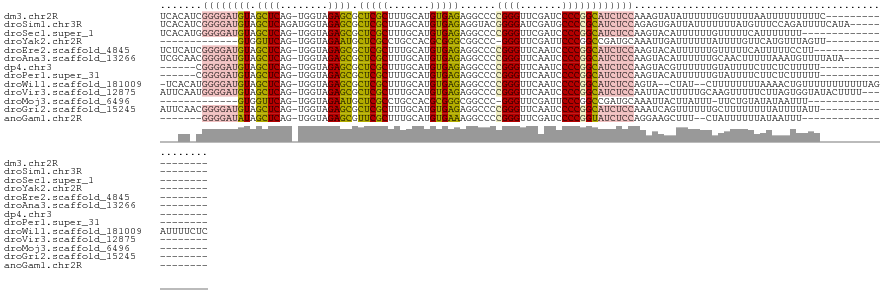
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,279,247 – 13,279,357 |
| Length | 110 |
| Max. P | 0.994205 |

| Location | 13,279,247 – 13,279,357 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 70.68 |
| Shannon entropy | 0.60240 |
| G+C content | 0.48113 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
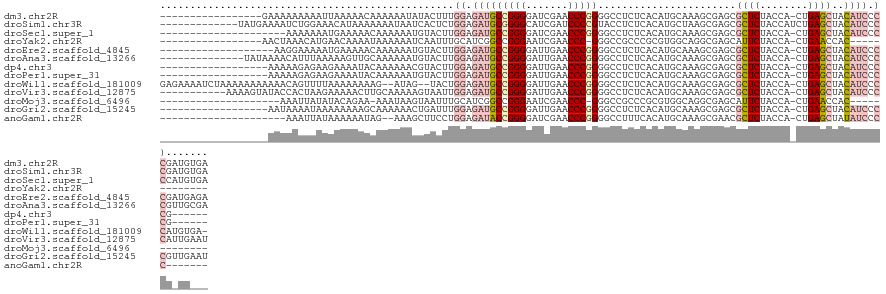
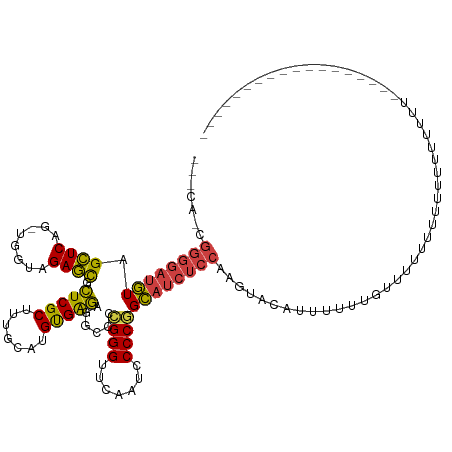
>dm3.chr2R 13279247 110 + 21146708 -----------------GAAAAAAAAAUUAAAAACAAAAAAUAUACUUUGGAGAUGCCGGGGAUCGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCGAUGUGA -----------------..........................(((.((((.(((((((((.......))))).....(((.(.......).))).((((.....-..))))..)))).)))).))). ( -26.70, z-score = -1.41, R) >droSim1.chr3R 8029824 115 + 27517382 -------------UAUGAAAAUCUGGAAACAUAAAAAAAUAAUCACUCUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGAUGUGA -------------..........((....))...................(((.(((((((.......))))))).)))(((((((((.....)))((((........))))..........)))))) ( -30.60, z-score = -2.19, R) >droSec1.super_1 10780311 106 + 14215200 ---------------------AAAAAAAUGAAAAACAAAAAAUGUACUUGGAGAUGCCGGGGAUCGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCCAUGUGA ---------------------.............................(((.(.(((((.......))))).).)))(((((((..........((((.....-..)))).........))))))) ( -26.31, z-score = -0.78, R) >droYak2.chr2R 12125935 96 + 21139217 -----------------AACUAAACAUGAACAAAAUAAAAAAUCAAUUUGCAUCGGCCGGGAAUCGAACCC-GGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCA-CUGAACCAC------------- -----------------...................................(((((((((.......)))-))((((((.((...)).)))).)).........-)))).....------------- ( -22.80, z-score = -0.30, R) >droEre2.scaffold_4845 10017252 108 - 22589142 -------------------AAGGAAAAAUGAAAAACAAAAAAUGUACUUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCGAUGAGA -------------------..(((..................((((..(((((.(.(((((.......))))).).))).))..))))........((((.....-..))))....)))......... ( -27.00, z-score = -0.67, R) >droAna3.scaffold_13266 6535159 113 - 19884421 --------------UAUAAAACAUUUAAAAAGUUGCAAAAAAUGUACUUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCGUUGCGA --------------..................((((((....((((..(((((.(.(((((.......))))).).))).))..))))........((((.....-..))))..........)))))) ( -29.00, z-score = -0.59, R) >dp4.chr3 16834602 103 + 19779522 ------------------AAAAAGAGAAGAAAAUACAAAAAACGUACUUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCG------ ------------------.........................(((..(((((.(.(((((.......))))).).))).))..))).........((((.....-..))))..........------ ( -23.60, z-score = -0.29, R) >droPer1.super_31 17621 103 - 935084 ------------------AAAAAGAGAAGAAAAUACAAAAAAUGUACUUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCG------ ------------------........................((((..(((((.(.(((((.......))))).).))).))..))))........((((.....-..))))..........------ ( -24.30, z-score = -0.51, R) >droWil1.scaffold_181009 3119524 122 - 3585778 GAGAAAAUCUAAAAAAAAAAACAGUUUUAAAAAAAAG--AUAG--UACUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCAUGUGA- .....................((((.(((........--.)))--.))))(((.(.(((((.......))))).).)))(((((((....(....)((((.....-..))))........)))))))- ( -28.60, z-score = -0.94, R) >droVir3.scaffold_12875 8015957 116 + 20611582 -----------AAAAGUAUACCACUAAGAAAAACUUGCAAAAAGUAAUUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCAUUGAAU -----------....(((...............(((((.....(((..(((((.(.(((((.......))))).).))).))..)))...))))).((((.....-..)))))))............. ( -27.50, z-score = -0.72, R) >droMoj3.scaffold_6496 16440248 92 + 26866924 --------------------AAAUUAUAUACAGAA-AAAUAAGUAAUUUGCAUCGGCCGGGAAUCGAACCC-GGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCA-CUGAACCAC------------- --------------------...............-......(((...(((.((((((.((........))-.))))(((.((...)).))))))))...)))..-.........------------- ( -24.00, z-score = -0.44, R) >droGri2.scaffold_15245 1508244 109 + 18325388 ------------------AAUAAAAUAAAAAAAAGCAAAAAACUGAUUUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCGUUGAAU ------------------...............................((.(((((((((.......))))).....(((.(.......).))).((((.....-..))))..)))).))....... ( -23.00, z-score = 0.16, R) >anoGam1.chr2R 51279363 97 - 62725911 ---------------------AAAUUAUAAAAAAUAG--AAAGCUUCCUGGAGAUACCGGGGAUCGAACCCGGGGCCUUUCACAUGCAAAGCGAACGCUCUACCA-CUGAGCUAUAUCCCC------- ---------------------................--..(((((..((((((..(((((.......))))).................((....))))).)))-..)))))........------- ( -23.10, z-score = -0.84, R) >consensus __________________AAAAAAAAAAAAAAAAACAAAAAAUGUACUUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCA_CUGAGCUACAUCCCCG_UG___ .................................................((.(((((((((.......))))).......................((((........))))..)))).))....... (-17.64 = -18.20 + 0.56)
| Location | 13,279,247 – 13,279,357 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 70.68 |
| Shannon entropy | 0.60240 |
| G+C content | 0.48113 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.78 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
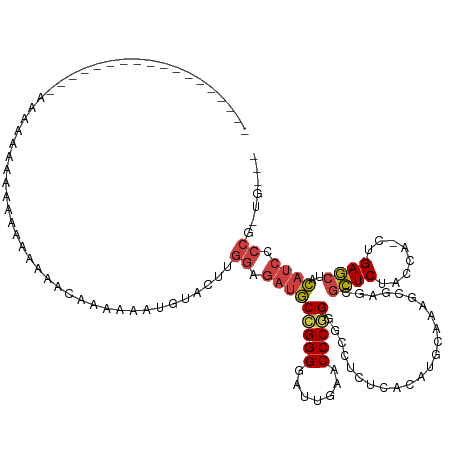
>dm3.chr2R 13279247 110 - 21146708 UCACAUCGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCGAUCCCCGGCAUCUCCAAAGUAUAUUUUUUGUUUUUAAUUUUUUUUUC----------------- .......(((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))................................----------------- ( -32.40, z-score = -1.67, R) >droSim1.chr3R 8029824 115 - 27517382 UCACAUCGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAGAGUGAUUAUUUUUUUAUGUUUCCAGAUUUUCAUA------------- ((((.((((((((((..((((.(((.(.((((....))))).))).))))......((((.......)))))))))))).)))))).............................------------- ( -37.20, z-score = -1.82, R) >droSec1.super_1 10780311 106 - 14215200 UCACAUGGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCGAUCCCCGGCAUCUCCAAGUACAUUUUUUGUUUUUCAUUUUUUU--------------------- .......(((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))............................--------------------- ( -32.10, z-score = -0.98, R) >droYak2.chr2R 12125935 96 - 21139217 -------------GUGGUUCAG-UGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCC-GGGUUCGAUUCCCGGCCGAUGCAAAUUGAUUUUUUAUUUUGUUCAUGUUUAGUU----------------- -------------...((((.(-((((((.........)))))))..))))((((.-(((.......)))))))((.(((((.(((....))).)))))))..........----------------- ( -30.90, z-score = -0.94, R) >droEre2.scaffold_4845 10017252 108 + 22589142 UCUCAUCGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAAGUACAUUUUUUGUUUUUCAUUUUUCCUU------------------- .......(((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))..............................------------------- ( -32.40, z-score = -1.49, R) >droAna3.scaffold_13266 6535159 113 + 19884421 UCGCAACGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAAGUACAUUUUUUGCAACUUUUUAAAUGUUUUAUA-------------- .......(((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))....((((((.............))))))......-------------- ( -33.42, z-score = -1.01, R) >dp4.chr3 16834602 103 - 19779522 ------CGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAAGUACGUUUUUUGUAUUUUCUUCUCUUUUU------------------ ------.(((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))(((((((.....)))))))............------------------ ( -35.60, z-score = -2.52, R) >droPer1.super_31 17621 103 + 935084 ------CGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAAGUACAUUUUUUGUAUUUUCUUCUCUUUUU------------------ ------.(((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))(((((((.....)))))))............------------------ ( -36.30, z-score = -2.87, R) >droWil1.scaffold_181009 3119524 122 + 3585778 -UCACAUGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAGUA--CUAU--CUUUUUUUUAAAACUGUUUUUUUUUUUAGAUUUUCUC -..((.((((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))))).--....--......(((((((..(....)..)))))))....... ( -33.60, z-score = -1.42, R) >droVir3.scaffold_12875 8015957 116 - 20611582 AUUCAAUGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAAUUACUUUUUGCAAGUUUUUCUUAGUGGUAUACUUUU----------- ......((((((((...((((.-..(((((((....)))))))...)))).....(((((.......)))))))))))))((((((.................))))))........----------- ( -34.53, z-score = -1.11, R) >droMoj3.scaffold_6496 16440248 92 - 26866924 -------------GUGGUUCAG-UGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCC-GGGUUCGAUUCCCGGCCGAUGCAAAUUACUUAUUU-UUCUGUAUAUAAUUU-------------------- -------------((((((..(-((((((.........)))))))(((..(((((.-(((.......)))))))).))).))))))......-...............-------------------- ( -31.50, z-score = -1.52, R) >droGri2.scaffold_15245 1508244 109 - 18325388 AUUCAACGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAAAUCAGUUUUUUGCUUUUUUUUAUUUUAUU------------------ .......(((((((...((((.-..(((((((....)))))))...)))).....(((((.......))))))))))))...............................------------------ ( -32.40, z-score = -1.52, R) >anoGam1.chr2R 51279363 97 + 62725911 -------GGGGAUAUAGCUCAG-UGGUAGAGCGUUCGCUUUGCAUGUGAAAGGCCCCGGGUUCGAUCCCCGGUAUCUCCAGGAAGCUUU--CUAUUUUUUAUAAUUU--------------------- -------(((((((..(((((.-..(((((((....)))))))...)))...)).(((((.......))))))))))))((((....))--))..............--------------------- ( -28.40, z-score = -0.62, R) >consensus ___CA_CGGGGAUGUAGCUCAG_UGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAAGUACAUUUUUUGUUUUUUUUUUUUUUUUU__________________ .......((((((((.((((........)))).(((((.......)))))......((((.......))))))))))))................................................. (-25.76 = -25.78 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:45 2011