
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,243,279 – 13,243,371 |
| Length | 92 |
| Max. P | 0.816399 |

| Location | 13,243,279 – 13,243,371 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Shannon entropy | 0.36930 |
| G+C content | 0.37986 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -9.83 |
| Energy contribution | -11.17 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
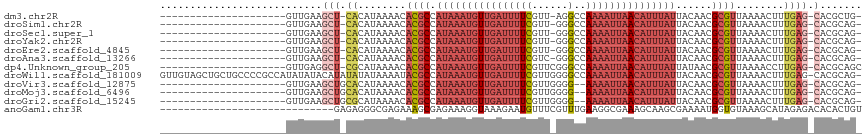
>dm3.chr2R 13243279 92 + 21146708 ---------------------GUUGAAGCU-CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUU-AGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCUG- ---------------------......(((-((........((((.(((((((((((((((.(..-...).)))))))))))))))......))))........))))-)......- ( -22.19, z-score = -3.05, R) >droSim1.chr2R 11977231 92 + 19596830 ---------------------GUUGAAGCU-CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUU-GGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((-((........((((.(((((((((((((((.(..-...).)))))))))))))))......))))........))))-)......- ( -22.19, z-score = -2.70, R) >droSec1.super_1 10744258 92 + 14215200 ---------------------GUUGAAGCU-CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUU-GGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((-((........((((.(((((((((((((((.(..-...).)))))))))))))))......))))........))))-)......- ( -22.19, z-score = -2.70, R) >droYak2.chr2R 2911233 92 - 21139217 ---------------------GUUGAAGCU-CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUU-GGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((-((........((((.(((((((((((((((.(..-...).)))))))))))))))......))))........))))-)......- ( -22.19, z-score = -2.70, R) >droEre2.scaffold_4845 9979122 92 - 22589142 ---------------------GUUGAAGCU-CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUU-GGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((-((........((((.(((((((((((((((.(..-...).)))))))))))))))......))))........))))-)......- ( -22.19, z-score = -2.70, R) >droAna3.scaffold_13266 6499079 92 - 19884421 ---------------------GUUGAAGCU-CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUC-GGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((-((........((((.(((((((((((((((.(..-...).)))))))))))))))......))))........))))-)......- ( -22.19, z-score = -2.84, R) >dp4.Unknown_group_205 12141 94 + 12543 ---------------------GUUGAGGCU-CGCAUAAAACACGCCAUAAAUGUUGAUUUUCGUUCGGGCCAAAAUUAACAUUUAUUAUAACGCGUUAAAACCUUGAG-CACGCAGC ---------------------((((..(((-((........((((.(((((((((((((((.((....)).)))))))))))))))......))))........))))-)...)))) ( -25.59, z-score = -2.63, R) >droWil1.scaffold_181009 3060987 115 - 3585778 GUUGUAGCUGCUGCCCCGCCAUAUAUACAUAUAUAUAAAAUACGCCAUAAAUGUUGAUUUUCGUUGGGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ((((((((.((......)).((((((....)))))).......)).(((((((((((((((.((....)).)))))))))))))))))))))((((............-.))))..- ( -25.32, z-score = -1.04, R) >droVir3.scaffold_12875 7964004 92 + 20611582 ---------------------GUUGAAGCUGCACAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGGG--AAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((((.((......((((.((((((((((((((((......)--)))))))))))))))......))))........)).)-)).))..- ( -24.04, z-score = -2.95, R) >droMoj3.scaffold_6496 7556764 92 - 26866924 ---------------------GUUGAAGCUGCACAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGGG--AAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((((.((......((((.((((((((((((((((......)--)))))))))))))))......))))........)).)-)).))..- ( -24.04, z-score = -2.95, R) >droGri2.scaffold_15245 1460327 92 + 18325388 ---------------------GUUGAAGCUGCGCAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGGG--AAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG-CACGCAG- ---------------------......(((((.((......((((.((((((((((((((((......)--)))))))))))))))......))))........)).)-)).))..- ( -24.54, z-score = -2.56, R) >anoGam1.chr3R 49087193 88 + 53272125 -----------------------------GAGAGGGCGAGAAAGCGAGAAAGGUAAAGAAUGUUUCGUUUGAAGGCGAAAGCAAGCGAAAAUGGUGUAAAGCAUAGAGACACACUGU -----------------------------......((......))...........((..((((((.((((...((....))...))))....(((.....))).))))))..)).. ( -16.50, z-score = -1.42, R) >consensus _____________________GUUGAAGCU_CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUU_GGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAG_CACGCAG_ ..............................................(((((((((((((((..........)))))))))))))))......((((..............))))... ( -9.83 = -11.17 + 1.33)

| Location | 13,243,279 – 13,243,371 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.93 |
| Shannon entropy | 0.36930 |
| G+C content | 0.37986 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -11.65 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
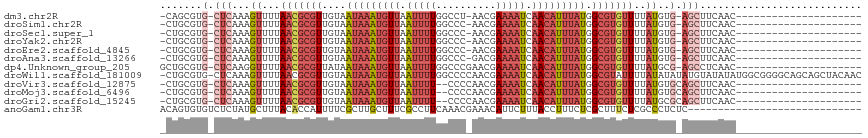
>dm3.chr2R 13243279 92 - 21146708 -CAGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCU-AACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUG-AGCUUCAAC--------------------- -......(-((((.......(((((((....(((((((((.(((((.(...-..).))))).))))))))).))))))).....))-)))......--------------------- ( -25.30, z-score = -2.71, R) >droSim1.chr2R 11977231 92 - 19596830 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCC-AACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUG-AGCUUCAAC--------------------- -......(-((((.......(((((((....(((((((((.(((((.(...-..).))))).))))))))).))))))).....))-)))......--------------------- ( -25.30, z-score = -2.62, R) >droSec1.super_1 10744258 92 - 14215200 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCC-AACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUG-AGCUUCAAC--------------------- -......(-((((.......(((((((....(((((((((.(((((.(...-..).))))).))))))))).))))))).....))-)))......--------------------- ( -25.30, z-score = -2.62, R) >droYak2.chr2R 2911233 92 + 21139217 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCC-AACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUG-AGCUUCAAC--------------------- -......(-((((.......(((((((....(((((((((.(((((.(...-..).))))).))))))))).))))))).....))-)))......--------------------- ( -25.30, z-score = -2.62, R) >droEre2.scaffold_4845 9979122 92 + 22589142 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCC-AACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUG-AGCUUCAAC--------------------- -......(-((((.......(((((((....(((((((((.(((((.(...-..).))))).))))))))).))))))).....))-)))......--------------------- ( -25.30, z-score = -2.62, R) >droAna3.scaffold_13266 6499079 92 + 19884421 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCC-GACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUG-AGCUUCAAC--------------------- -......(-((((.......(((((((....(((((((((.(((((.(...-..).))))).))))))))).))))))).....))-)))......--------------------- ( -25.30, z-score = -2.40, R) >dp4.Unknown_group_205 12141 94 - 12543 GCUGCGUG-CUCAAGGUUUUAACGCGUUAUAAUAAAUGUUAAUUUUGGCCCGAACGAAAAUCAACAUUUAUGGCGUGUUUUAUGCG-AGCCUCAAC--------------------- ..((.(.(-(((...((...((((((((((((...(((((.(((((.(......).))))).)))))))))))))))))....)))-)))).))..--------------------- ( -24.10, z-score = -1.32, R) >droWil1.scaffold_181009 3060987 115 + 3585778 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCCCAACGAAAAUCAACAUUUAUGGCGUAUUUUAUAUAUAUGUAUAUAUGGCGGGGCAGCAGCUACAAC -((((.((-(((...((....)).(((..(((((((((((.(((((.(......).))))).))))))))).((((((.......))))))...))..))))))))))))....... ( -30.50, z-score = -1.57, R) >droVir3.scaffold_12875 7964004 92 - 20611582 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUU--CCCCAACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUGCAGCUUCAAC--------------------- -..((.((-(.((.......(((((((....(((((((((.(((((--(......)))))).))))))))).)))))))...)).)))))......--------------------- ( -24.40, z-score = -3.06, R) >droMoj3.scaffold_6496 7556764 92 + 26866924 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUU--CCCCAACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUGCAGCUUCAAC--------------------- -..((.((-(.((.......(((((((....(((((((((.(((((--(......)))))).))))))))).)))))))...)).)))))......--------------------- ( -24.40, z-score = -3.06, R) >droGri2.scaffold_15245 1460327 92 - 18325388 -CUGCGUG-CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUU--CCCCAACGAAAAUCAACAUUUAUGGCGUGUUUUAUGCGCAGCUUCAAC--------------------- -(((((((-(.....))...(((((((....(((((((((.(((((--(......)))))).))))))))).)))))))....)))))).......--------------------- ( -25.70, z-score = -3.32, R) >anoGam1.chr3R 49087193 88 - 53272125 ACAGUGUGUCUCUAUGCUUUACACCAUUUUCGCUUGCUUUCGCCUUCAAACGAAACAUUCUUUACCUUUCUCGCUUUCUCGCCCUCUC----------------------------- ...(((((...........))))).......(..((.(((((........)))))))..)............((......))......----------------------------- ( -6.80, z-score = -0.33, R) >consensus _CUGCGUG_CUCAAAGUUUUAACGCGUUGUAAUAAAUGUUAAUUUUGGCCC_AACGAAAAUCAACAUUUAUGGCGUGUUUUAUGUG_AGCUUCAAC_____________________ ...(((((............(((((((....(((((((((.(((((..........))))).))))))))).))))))).)))))................................ (-11.65 = -12.57 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:41 2011