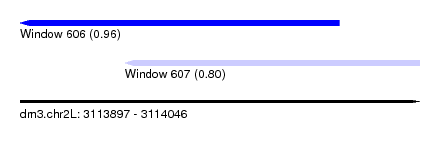
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,113,897 – 3,114,046 |
| Length | 149 |
| Max. P | 0.964486 |

| Location | 3,113,897 – 3,114,016 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.58710 |
| G+C content | 0.46151 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
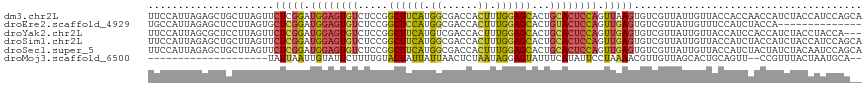
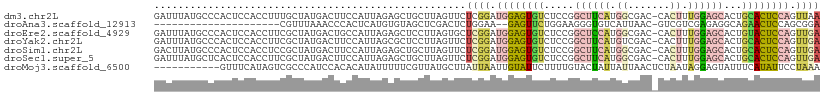
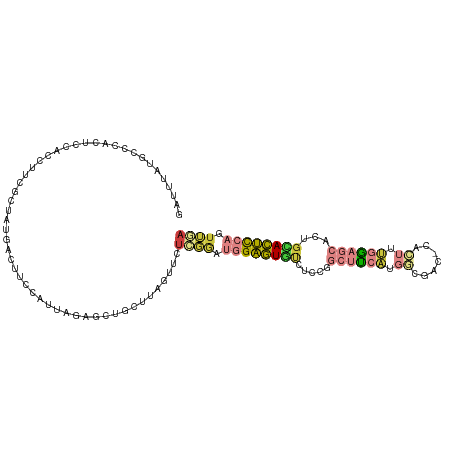
>dm3.chr2L 3113897 119 - 23011544 UUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGACCACUUUGGAGCACUGCACUCCAGUUAAGUGUCGUUAUUGUUACCACCAACCAUCUACCAUCCAGCA .......((((((....)))))).((((((((((.....((...(((((((((.(((((((((.......)))))...))))))))))).))...))......)).)).)))))).... ( -33.20, z-score = -1.33, R) >droEre2.scaffold_4929 3157118 105 - 26641161 UGCCAUUAGAGCUCCUUAGUGCUCGGAUGGAGUGUCUCCGGCUCCAUGGCGACCACUUUGGAGCACUGUACUCCAGUUGAGUGUCGUUAUUGUUUCCAUCUACCA-------------- ......(((((((((..((((.(((.(((((((.......)))))))..))).))))..)))))((..(((((.....)))))..))...........))))...-------------- ( -34.10, z-score = -1.57, R) >droYak2.chr2L 3105985 116 - 22324452 UUCCAUUAGCGCUCCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGUCGACCACUUUGGAGCACUGCACUCCAGUUGAGUGUCGUUAUUGUUACCAUCCACCAUCUACCUACCA--- ...((.(((((((((..(((..(((((((((((.......))))))).))))..)))..)))))...((((((.....)))))).)))).))........................--- ( -33.40, z-score = -2.33, R) >droSim1.chr2L 3052407 119 - 22036055 UUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGACCACUUUGGAGCACUGCACUCCAGUUGAGUGUCGUUAUUGUUACCAUCUACCAUCUACCAUCCAGCA .......((((((....)))))).((((((((((.....((...(((((((((.(((((((((.......)))))...))))))))))).))...))......)).)).)))))).... ( -33.60, z-score = -0.94, R) >droSec1.super_5 1265916 119 - 5866729 UUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGACCACUUUGGAGCACUGCACUCCAGUUGAGUGUCGUUAUUGUUACCAUCUACUAUCUACAAUCCAGCA .......((((((....)))))).((((((((...))))((...(((((((((.(((((((((.......)))))...))))))))))).))...))..............)))).... ( -31.80, z-score = -0.44, R) >droMoj3.scaffold_6500 23165559 95 - 32352404 --------------------UAUUAAUUGUAUUCUUUUGUACUAUUAUUAACUCUAAUAGGAGUAUUUCAUAUUCCUAAAACGUUGUUAGCACUGCAGUU--CCGUUUACUAAUGCA-- --------------------((.((((.((((......)))).)))).)).......(((((((((...)))))))))(((((((((.......))))..--.))))).........-- ( -14.10, z-score = -0.75, R) >consensus UUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGACCACUUUGGAGCACUGCACUCCAGUUGAGUGUCGUUAUUGUUACCAUCUACCAUCUACCAUCCAG__ .....................(((((.((((((((.....((((((.((......)).))))))...)))))))).)))))...................................... (-18.96 = -18.88 + -0.08)
| Location | 3,113,936 – 3,114,046 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 62.91 |
| Shannon entropy | 0.74336 |
| G+C content | 0.48532 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.75 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
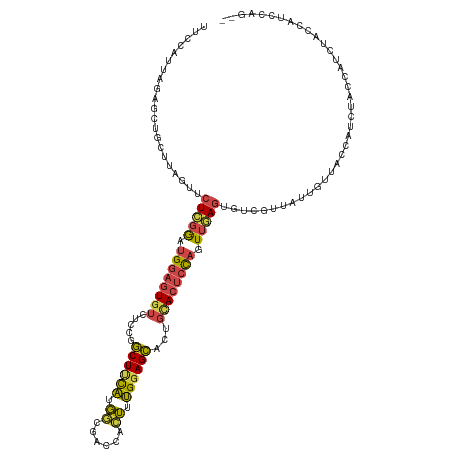
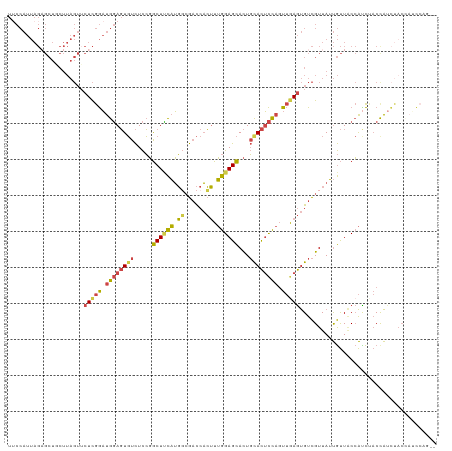
>dm3.chr2L 3113936 110 - 23011544 GAUUUAUGCCCACUCCACCUUUGCUAUGACUUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGAC-CACUUUGGAGCACUGCACUCCAGUUAA ..........................((((.(((...((((((....)))))).)))((((((((.....((((((.((....-..)).))))))...)))))))))))). ( -29.70, z-score = -0.83, R) >droAna3.scaffold_12913 332950 87 - 441482 ---------------------CGUUUAAACCCACUCAUGUGUAGCUCGACUCUGGAA--GAGUUCUGGAAGGGUGUCAUUAAC-GUCGUCGAGAGGCAGAACUCCAGCGGA ---------------------.........((.((..(.(((..((((((((..((.--....))..))..(..((.....))-..)))))))..))).).....)).)). ( -20.80, z-score = 1.19, R) >droEre2.scaffold_4929 3157143 110 - 26641161 GAUUUAUGCCCACUCCACCUUCGCUAUGACUGCCAUUAGAGCUCCUUAGUGCUCGGAUGGAGUGUCUCCGGCUCCAUGGCGAC-CACUUUGGAGCACUGUACUCCAGUUGA ............(((((...((((((((...(((....((((........))))....((((...)))))))..)))))))).-.....))))).((((.....))))... ( -34.20, z-score = -1.41, R) >droYak2.chr2L 3106021 110 - 22324452 GAUUUAUGCCCACUCCACCUUCGCUAUGACUUCCAUUAGCGCUCCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGUCGAC-CACUUUGGAGCACUGCACUCCAGUUGA ......(((.............((((((.....)).))))(((((..(((..(((((((((((.......))))))).)))).-.)))..)))))...))).......... ( -29.60, z-score = -1.27, R) >droSim1.chr2L 3052446 110 - 22036055 GACUUAUGCCCACUCCACCUCCGCUAUGACUUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGAC-CACUUUGGAGCACUGCACUCCAGUUGA ((((..(((...(((((....((((((((.(((((((((((((....))))))..))))))).((.....)).))))))))..-.....)))))....)))....)))).. ( -30.70, z-score = -0.59, R) >droSec1.super_5 1265955 110 - 5866729 GAUUUAUGCUCACUCCACCUUCGCUAUGACUUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGAC-CACUUUGGAGCACUGCACUCCAGUUGA ......(((...(((((...(((((((((.(((((((((((((....))))))..))))))).((.....)).))))))))).-.....)))))....))).......... ( -32.20, z-score = -1.14, R) >droMoj3.scaffold_6500 23165594 100 - 32352404 -----------GUUUCAUAGUCGCCCAUCCACACAUAUUUUUCGUUAUGCUUAUUAAUUGUAUUCUUUUGUACUAUUAUUAACUCUAAUAGGAGUAUUUCAUAUUCCUAAA -----------......(((.............((((........)))).(((.((((.((((......)))).)))).)))..))).(((((((((...))))))))).. ( -12.50, z-score = -1.91, R) >consensus GAUUUAUGCCCACUCCACCUUCGCUAUGACUUCCAUUAGAGCUGCUUAGUUCUCGGAUGGAGUGUCUCCGGCUUCAUGGCGAC_CACUUUGGAGCACUGCACUCCAGUUGA ....................................................((((.((((((((.....((((((.............))))))...)))))))).)))) (-11.80 = -12.75 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:05 2011