| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,170,933 – 13,171,033 |
| Length | 100 |
| Max. P | 0.540162 |

| Location | 13,170,933 – 13,171,033 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.02 |
| Shannon entropy | 0.48724 |
| G+C content | 0.50364 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -14.44 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
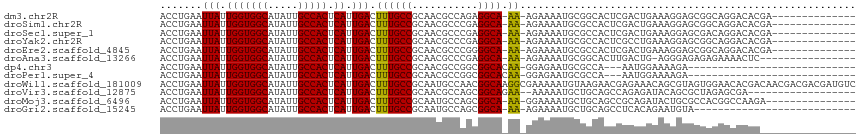
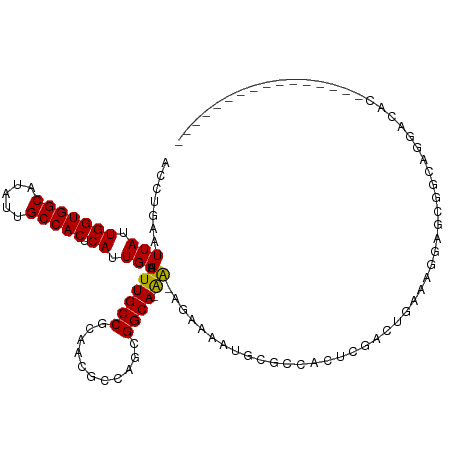
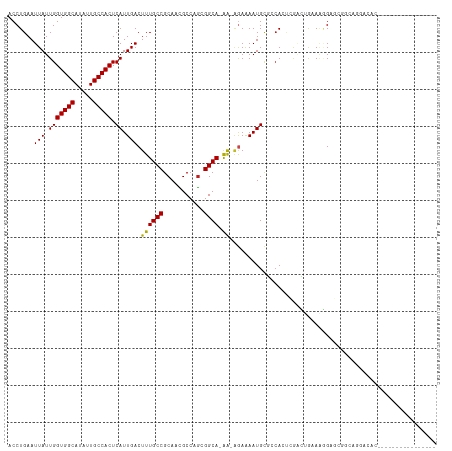
>dm3.chr2R 13170933 100 - 21146708 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCAGAGGCA-AA-AGAAAAUGCGGCACUCGACUGAAAGGAGCGGCAGGACACGA-------------- .(((...(((.(((((((.....))))).)).)))....(((((...(((....(((-..-......))))))......((....)).))))))))......-------------- ( -30.80, z-score = -1.79, R) >droSim1.chr2R 11898906 100 - 19596830 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCCGAGGCA-AA-AGAAAAUGCGCCACUCGACUGAAAGGAGCGGCAGGACACGA-------------- .(((...(((.(((((((.....))))).)).)))....(((((......(((((((-..-......)))....)))).((....)).))))))))......-------------- ( -30.40, z-score = -1.66, R) >droSec1.super_1 10672239 100 - 14215200 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCCGAGGCA-AA-AGAAAAUGCGCCACUCGACUGAAAGGAGCGACAGGACACGA-------------- .((((.......((((((.....))))))((((..(((((((.(.......).))).-))-))..))))(((..((........))..))).))))......-------------- ( -27.40, z-score = -1.47, R) >droYak2.chr2R 2836420 100 + 21139217 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCCGAGGCA-AA-AGAAAAUGCGCCACUCGCCUGAAAGGAGCGGCAGGACACGA-------------- .(((...(((.(((((((.....))))).)).)))....(((((......(((((((-..-......)))....)))).((....)).))))))))......-------------- ( -29.50, z-score = -0.85, R) >droEre2.scaffold_4845 9904407 100 + 22589142 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCCGGGGCA-AA-AGAAAAUGCGCCACUCGACUGAAAGGAGCGGCAGGACACGA-------------- .(((...(((.(((((((.....))))).)).)))....(((((.......((.(((-..-......))).))......((....)).))))))))......-------------- ( -32.70, z-score = -1.81, R) >droAna3.scaffold_13266 6430011 97 + 19884421 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCCGAGGCA-AA-AGAAAAUGCGGCACUUGACUG-AGGGAGAGAGAAAACUC---------------- .(((.(.(((.(((((((.....))))).)).)))...(((((((..(((...))).-..-......)))))))......).-)))..(((......)))---------------- ( -25.60, z-score = -1.32, R) >dp4.chr3 16736071 84 - 19779522 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCGGCGGCACAA-GGAGAAUGCGCCA---AAUGGAAAAGA---------------------------- .(((...(((.(((((((.....))))).)).)))...((((((.......))))))..)-))........((.---...))......---------------------------- ( -24.20, z-score = -0.81, R) >droPer1.super_4 7128639 84 + 7162766 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCGGCGGCACAA-GGAGAAUGCGCCA---AAUGGAAAAGA---------------------------- .(((...(((.(((((((.....))))).)).)))...((((((.......))))))..)-))........((.---...))......---------------------------- ( -24.20, z-score = -0.81, R) >droWil1.scaffold_181009 2948195 116 + 3585778 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAAUGCCAACGGCAAGGCGAAAAAUGUAAGAACGAGAAACAGCGUAGUGGAACACGACAACGACGACGAUGUC (((((.......((((((.....))))))..(((.((((((((.........)))))))))))...................))).)).(((...)))((((.((....)).)))) ( -28.90, z-score = -0.88, R) >droVir3.scaffold_12875 7866446 96 - 20611582 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCAGCGGCAGAA--AAAAAUGCUGCAGCCAGAGAUACAGCGCUAGAGCGA------------------ ..(((.(((.((((((((.....)))...((((...((((((((.......)))))))).--...)))).....))))).))).)))(((....))).------------------ ( -30.40, z-score = -1.59, R) >droMoj3.scaffold_6496 7432772 99 + 26866924 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAAUGCCAGCGGCA-AA-GGAAAAUGCUGCAGCCGCAGAUACUGCGCCACGGCCAAGA--------------- .......(((.(((((((.....))))).)).)))(((((((((.......))))))-))-)......((((..((.((((...))))))..)))).....--------------- ( -36.70, z-score = -2.20, R) >droGri2.scaffold_15245 1365855 87 - 18325388 ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAAUGCCAGCGGCA-AA-AGAAAAUGCUGCAGCCUCACAGAAUGUA--------------------------- ..(((.......((((((.....))))))((((..(((((((((.......))))).-))-))..))))...........)))......--------------------------- ( -25.50, z-score = -1.28, R) >consensus ACCUGAAUUAUUGGUGGCAUAUUGCCACUCAUUGACUUUGCCGCAACGCCAGCGGCA_AA_AGAAAAUGCGCCACUCGACUGAAAGGAGCGGCAGGACAC________________ ............((((((.....))))))((((..((.((((...........))))....))..))))............................................... (-14.44 = -14.50 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:33 2011