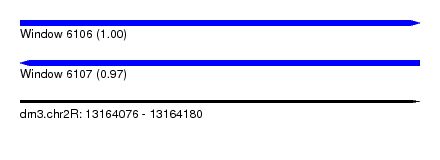
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,164,076 – 13,164,180 |
| Length | 104 |
| Max. P | 0.999287 |

| Location | 13,164,076 – 13,164,180 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 60.98 |
| Shannon entropy | 0.75251 |
| G+C content | 0.28342 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -7.60 |
| Energy contribution | -10.16 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
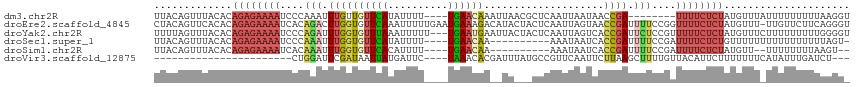
>dm3.chr2R 13164076 104 + 21146708 ACCUUAAAAAAAAAUAAACAUAGAGAAAA--------UCGGUUAUUAAUUGAGCGUUAAUUUGUUCA----AAAAUAUGAACAACAAAUUUGGGAUUUUCUCUGUGUAAACUGUAA .................((((((((((((--------(((.(((.....))).)......(((((((----......))))))).........))))))))))))))......... ( -26.00, z-score = -4.68, R) >droEre2.scaffold_4845 9897807 115 - 22589142 ACCCUGAAGAACAA-AAACAUAGAGAAAACCGGAAAAUCGGUUACUAAUUGAGUAGUAUGUCUUUCAUUCAAAAAUUUGAACACCAAGUCUGUGAUUUUCUCUGUGUGAACUGUAG ..........(((.-..(((((((((((((((((.....((((((((......))))).........(((((....))))).)))...)))).).))))))))))))....))).. ( -25.30, z-score = -1.68, R) >droYak2.chr2R 2829833 113 - 21139217 ACCCCAAAAAAAAAGAAACAUAGAGAAAAACGGAGAAUCGGUGACUAAUUGAGUAGUAAUUCAUUCA---AAAAAUUUAAACACCAAAUCUGGGAUUUUCUCUGUGUAAACUAAAA .................((((((((((((.((((.....((((..(((((((((........)))))---).....)))..))))...))))...))))))))))))......... ( -24.50, z-score = -3.02, R) >droSec1.super_1 10665831 101 + 14215200 -ACUAAAAAAAAAAAAAAAACAGAGAAAAUCGGAAAAUCGGUGAUUAUUU----------UUGUUCA----AAAAUAUGAACACCAAAUUUGGGAUUUUCUCUGUGUAAACUGUAA -..................((((((((((((..(((...((((.((((.(----------((.....----.))).)))).))))...)))..))))))))))))........... ( -24.20, z-score = -4.26, R) >droSim1.chr2R 11892489 98 + 19596830 --ACUUAAAAAAAA--AACAUAGAGAAAAUCGGAAAAUCGGUGAUUAUUU----------UUGUUCA----AAAAUGUGAACACCAAAUUUGUGAUUUUCUCUGUGUAAACUGUAA --............--.(((((((((((((((.(((...((((.((((.(----------((.....----.))).)))).))))...))).)))))))))))))))......... ( -29.60, z-score = -6.03, R) >droVir3.scaffold_12875 7859069 86 + 20611582 ---AGAUCAAAUAUGAAAAAAAGAAUGUAACAAAAGCUUAAGAAUUGAACGGCAUAAAUCGUGUUUA----GAAUCAUAAUUAUCGAAUCCAG----------------------- ---.((((((((((((......(.......)....(((............))).....)))))))).----).))).................----------------------- ( -5.60, z-score = 1.39, R) >consensus _CCCUAAAAAAAAA_AAACAUAGAGAAAAACGGAAAAUCGGUGAUUAAUUGAG___UA__UUGUUCA____AAAAUAUGAACACCAAAUCUGGGAUUUUCUCUGUGUAAACUGUAA .................((((((((((((..........((((.((((............................)))).))))..........))))))))))))......... ( -7.60 = -10.16 + 2.56)

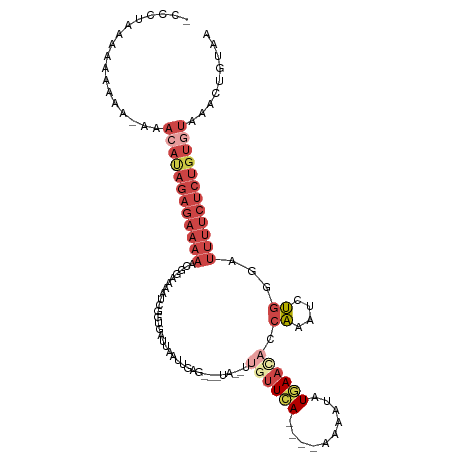
| Location | 13,164,076 – 13,164,180 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 60.98 |
| Shannon entropy | 0.75251 |
| G+C content | 0.28342 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -5.09 |
| Energy contribution | -7.01 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13164076 104 - 21146708 UUACAGUUUACACAGAGAAAAUCCCAAAUUUGUUGUUCAUAUUUU----UGAACAAAUUAACGCUCAAUUAAUAACCGA--------UUUUCUCUAUGUUUAUUUUUUUUUAAGGU ....(((..(((.((((((((((.........(((((((......----)))))))(((((.......)))))....))--------)))))))).)))..)))............ ( -20.80, z-score = -3.56, R) >droEre2.scaffold_4845 9897807 115 + 22589142 CUACAGUUCACACAGAGAAAAUCACAGACUUGGUGUUCAAAUUUUUGAAUGAAAGACAUACUACUCAAUUAGUAACCGAUUUUCCGGUUUUCUCUAUGUUU-UUGUUCUUCAGGGU ..((((...(((.((((((((((..(((.(((((((((((....))))))........(((((......)))))))))).)))..)))))))))).)))..-)))).......... ( -25.90, z-score = -1.40, R) >droYak2.chr2R 2829833 113 + 21139217 UUUUAGUUUACACAGAGAAAAUCCCAGAUUUGGUGUUUAAAUUUUU---UGAAUGAAUUACUACUCAAUUAGUCACCGAUUCUCCGUUUUUCUCUAUGUUUCUUUUUUUUUGGGGU ....((...(((.((((((((....(((.((((((.((((.....(---(((.((......)).)))))))).)))))).)))....)))))))).)))..))............. ( -21.70, z-score = -1.18, R) >droSec1.super_1 10665831 101 - 14215200 UUACAGUUUACACAGAGAAAAUCCCAAAUUUGGUGUUCAUAUUUU----UGAACAA----------AAAUAAUCACCGAUUUUCCGAUUUUCUCUGUUUUUUUUUUUUUUUUAGU- ...........((((((((((((..(((.((((((....((((((----(....))----------)))))..)))))).)))..))))))))))))..................- ( -26.70, z-score = -5.90, R) >droSim1.chr2R 11892489 98 - 19596830 UUACAGUUUACACAGAGAAAAUCACAAAUUUGGUGUUCACAUUUU----UGAACAA----------AAAUAAUCACCGAUUUUCCGAUUUUCUCUAUGUU--UUUUUUUUAAGU-- .........(((.((((((((((..(((.((((((.....(((((----(....))----------))))...)))))).)))..)))))))))).))).--............-- ( -23.10, z-score = -4.27, R) >droVir3.scaffold_12875 7859069 86 - 20611582 -----------------------CUGGAUUCGAUAAUUAUGAUUC----UAAACACGAUUUAUGCCGUUCAAUUCUUAAGCUUUUGUUACAUUCUUUUUUUCAUAUUUGAUCU--- -----------------------..((((.(((....(((((...----.....(((........)))..........(((....)))............))))).)))))))--- ( -6.30, z-score = 1.21, R) >consensus UUACAGUUUACACAGAGAAAAUCCCAAAUUUGGUGUUCAUAUUUU____UGAACAA__UA___CUCAAUUAAUCACCGAUUUUCCGAUUUUCUCUAUGUUU_UUUUUUUUUAGGG_ .............((((((((....(((.((((((......................................)))))).)))....))))))))..................... ( -5.09 = -7.01 + 1.92)
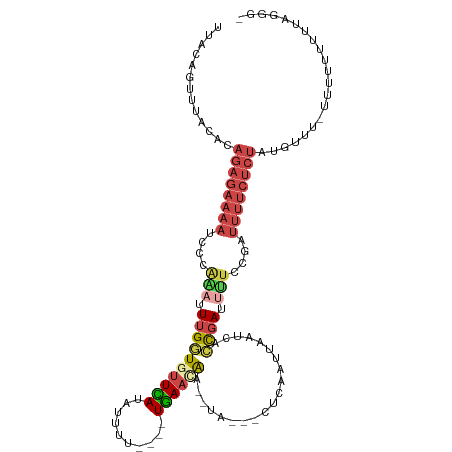
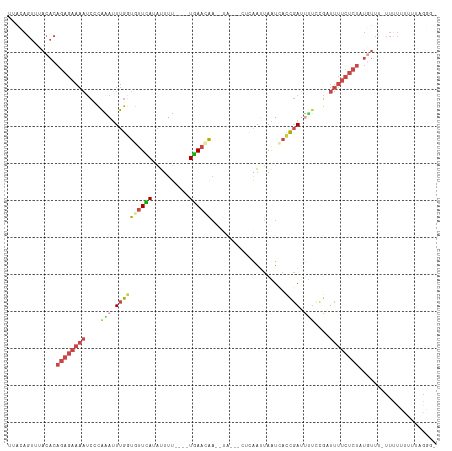
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:32 2011