| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,155,700 – 13,155,780 |
| Length | 80 |
| Max. P | 0.970286 |

| Location | 13,155,700 – 13,155,780 |
|---|---|
| Length | 80 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.39255 |
| G+C content | 0.41570 |
| Mean single sequence MFE | -18.93 |
| Consensus MFE | -10.62 |
| Energy contribution | -10.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
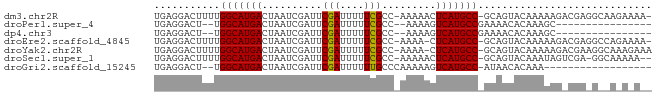
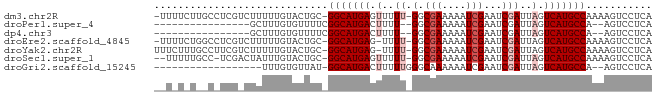
>dm3.chr2R 13155700 80 + 21146708 UGAGGACUUUUGGCAUGACUAAUCGAUUCGAUUUUUCGCC-AAAAACUCAUGCC-GCAGUACAAAAAGACGAGGCAAGAAAA- ((((...(((((((..((..(((((...)))))..)))))-)))).))))((((-...((........))..))))......- ( -21.50, z-score = -2.64, R) >droPer1.super_4 7116667 63 - 7162766 UGAGGACU--UGGCAUGACUAAUCGAUUCGAUUUUUCGCC--AAAAGUCAUGCCGAAAACACAAAGC---------------- .......(--((((((((((...(((.........)))..--...)))))))))))...........---------------- ( -17.30, z-score = -2.77, R) >dp4.chr3 16724442 63 + 19779522 UGAGGACU--UGGCAUGACUAAUCGAUUCGAUUUUUCGCC--AAAAGUCAUGCCGAAAACACAAAGC---------------- .......(--((((((((((...(((.........)))..--...)))))))))))...........---------------- ( -17.30, z-score = -2.77, R) >droEre2.scaffold_4845 9889401 79 - 22589142 UGAGGACUUUUGGCAUGACUAAUCGAUUCGAUUUUUCGCC-AAAA-CUCAUGCC-GCAGUACAAAAAGACGAGGCCAGAAAA- ((((...(((((((..((..(((((...)))))..)))))-))))-)))).(((-...((........))..))).......- ( -18.20, z-score = -1.36, R) >droYak2.chr2R 2820510 80 - 21139217 UGAGGACUUUUGGCAUGACUAAUCGAUUCGAUUUUUCGCC-AAAA-CUCAUGCC-GCAGUACAAAAAGACGAAGGCAAAGAAA ((((...(((((((..((..(((((...)))))..)))))-))))-))))((((-...((........))...))))...... ( -20.40, z-score = -2.24, R) >droSec1.super_1 10657508 78 + 14215200 UGAGGACUUUUGGCAUGACUAAUCGAUUCGAUUUUUCGCC-AAAAACUCAUGCC-GCAGUACAAAUAGUCGA-GGCAAAAA-- ((((...(((((((..((..(((((...)))))..)))))-)))).))))((((-...(.((.....)))..-))))....-- ( -21.00, z-score = -2.41, R) >droGri2.scaffold_15245 1347010 62 + 18325388 UGAGGACU--UGGCAUGACUAAUCGAUUCGAUUUUUUGCCCAAAAAGUCAUGCC-AUAACACAAA------------------ ((.(....--(((((((((((((((...)))))((((....)))))))))))))-)...).))..------------------ ( -16.80, z-score = -3.34, R) >consensus UGAGGACUUUUGGCAUGACUAAUCGAUUCGAUUUUUCGCC_AAAAACUCAUGCC_GCAGUACAAAAAG_CGA_G__A_A_A__ ...........(((((((..........(((....))).........)))))))............................. (-10.62 = -10.50 + -0.12)
| Location | 13,155,700 – 13,155,780 |
|---|---|
| Length | 80 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.39255 |
| G+C content | 0.41570 |
| Mean single sequence MFE | -18.90 |
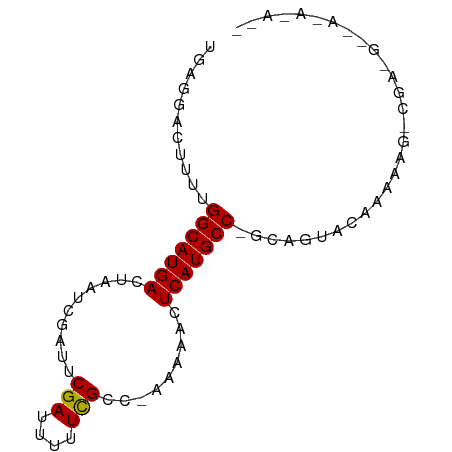
| Consensus MFE | -11.57 |
| Energy contribution | -11.71 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
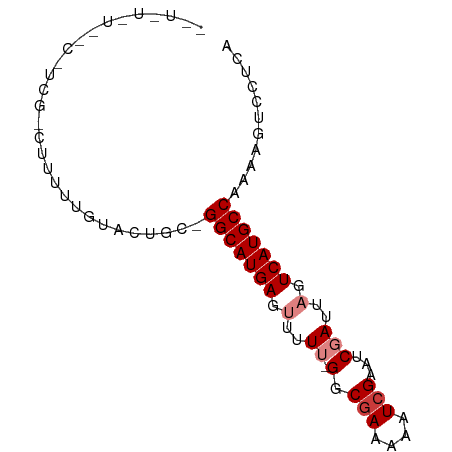
>dm3.chr2R 13155700 80 - 21146708 -UUUUCUUGCCUCGUCUUUUUGUACUGC-GGCAUGAGUUUUU-GGCGAAAAAUCGAAUCGAUUAGUCAUGCCAAAAGUCCUCA -......((((.((((.....).)).).-))))(((((((((-(((((..(((((...)))))..))..))))))))..)))) ( -19.30, z-score = -1.57, R) >droPer1.super_4 7116667 63 + 7162766 ----------------GCUUUGUGUUUUCGGCAUGACUUUU--GGCGAAAAAUCGAAUCGAUUAGUCAUGCCA--AGUCCUCA ----------------.............(((((((((.((--(.(((....)))...)))..))))))))).--........ ( -18.60, z-score = -2.47, R) >dp4.chr3 16724442 63 - 19779522 ----------------GCUUUGUGUUUUCGGCAUGACUUUU--GGCGAAAAAUCGAAUCGAUUAGUCAUGCCA--AGUCCUCA ----------------.............(((((((((.((--(.(((....)))...)))..))))))))).--........ ( -18.60, z-score = -2.47, R) >droEre2.scaffold_4845 9889401 79 + 22589142 -UUUUCUGGCCUCGUCUUUUUGUACUGC-GGCAUGAG-UUUU-GGCGAAAAAUCGAAUCGAUUAGUCAUGCCAAAAGUCCUCA -.......(((.((((.....).)).).-)))..((.-((((-(((((..(((((...)))))..))..))))))).)).... ( -18.50, z-score = -0.92, R) >droYak2.chr2R 2820510 80 + 21139217 UUUCUUUGCCUUCGUCUUUUUGUACUGC-GGCAUGAG-UUUU-GGCGAAAAAUCGAAUCGAUUAGUCAUGCCAAAAGUCCUCA ......((((...(((.....).))...-)))).((.-((((-(((((..(((((...)))))..))..))))))).)).... ( -18.60, z-score = -1.31, R) >droSec1.super_1 10657508 78 - 14215200 --UUUUUGCC-UCGACUAUUUGUACUGC-GGCAUGAGUUUUU-GGCGAAAAAUCGAAUCGAUUAGUCAUGCCAAAAGUCCUCA --....((((-.(.((.....))...).-))))(((((((((-(((((..(((((...)))))..))..))))))))..)))) ( -19.80, z-score = -1.71, R) >droGri2.scaffold_15245 1347010 62 - 18325388 ------------------UUUGUGUUAU-GGCAUGACUUUUUGGGCAAAAAAUCGAAUCGAUUAGUCAUGCCA--AGUCCUCA ------------------.........(-(((((((((..((((.............))))..))))))))))--........ ( -18.92, z-score = -3.12, R) >consensus __U_U_U__C_UCG_CUUUUUGUACUGC_GGCAUGAGUUUUU_GGCGAAAAAUCGAAUCGAUUAGUCAUGCCAAAAGUCCUCA .............................(((((((.........(((....)))..........)))))))........... (-11.57 = -11.71 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:30 2011