
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,111,735 – 13,111,873 |
| Length | 138 |
| Max. P | 0.989473 |

| Location | 13,111,735 – 13,111,835 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.43 |
| Shannon entropy | 0.59228 |
| G+C content | 0.52585 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -10.94 |
| Energy contribution | -10.76 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
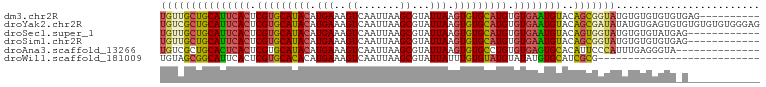
>dm3.chr2R 13111735 100 + 21146708 ----CCUUUCGUUCGCCACCCG-CCGUCCCGC--------AUCCUCAUCGUUCGGCUGUCAUGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUG-- ----.(((.....(((.....(-(......))--------.............((((.((((((...(((((......))))).)))))).))))......)))....)))..-- ( -21.40, z-score = -1.38, R) >droEre2.scaffold_4845 9847275 99 - 22589142 ----CCUUUCGC-CGCC-CCCCGCCGUCCCGC--------AUCCUCAUCGUUCGGCUGUCAUGUCGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUG-- ----.(((....-(((.-....((......))--------.............((((.((((((...(((((......))))).)))))).))))......)))....)))..-- ( -20.60, z-score = -1.32, R) >droYak2.chr2R 2780179 108 - 21139217 ----CCUUUCGCUCGCCACUCC-CCGCCCCUCCGUCACGGGUCCUCAUCGUUCGGCUGUCAUGUCGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUG-- ----.(((.((((........(-(((...........))))............((((.((((((...(((((......))))).)))))).)))).....))))....)))..-- ( -24.80, z-score = -1.57, R) >droSec1.super_1 10615248 101 + 14215200 ----CCUUUCGUUCGCCACCCCGCCGUCCCGG--------AUCCUCAUCGUUCGGCUGUCAUGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUG-- ----.(((.....(((....(((......)))--------.............((((.((((((...(((((......))))).)))))).))))......)))....)))..-- ( -22.90, z-score = -1.36, R) >droSim1.chr2R 11844002 100 + 19596830 ----CCUUUCGCUCGC-ACCCCGCCGUCCCGG--------AUCCUCAUCGUUCGGCUGUCAUGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUG-- ----.(((.....(((-...(((......)))--------.............((((.((((((...(((((......))))).)))))).))))......)))....)))..-- ( -23.80, z-score = -1.39, R) >droPer1.super_4 7081217 99 - 7162766 ----CCUCUCGGCCCU--CUCGACCGGCGAGC--------AUGAUGCUCCUUUUGCUGUCAUGUAGCUGCAUUCAGUCGUGCAUGCAUGAAAGUCAAUUAAGCGUAUUAAGUG-- ----.((((((((...--...).)))).))).--------...(((((....(((((.(((((((..(((((......)))))))))))).)).)))...)))))........-- ( -25.40, z-score = -0.13, R) >droAna3.scaffold_13266 6377672 105 - 19884421 CGGGAGUCCGGACCUGCCCCCUACCCUCUCCC--------AAUCUUCACUUCGCGCUGUCAUGUCGCUGCACUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUG-- .(((((...((............))..)))))--------......(((((.(((((.((((((...(((((......))))).))))))..........)))))...)))))-- ( -29.70, z-score = -3.06, R) >droWil1.scaffold_181009 2870703 91 - 3585778 -----------CUUUUCAUUCCUCCCUUGCCC-----------UUAACUUUUGGGCCAAAGUGUAGCGGCAUUCACUCGUGCACACAUGAAAGUCAAUUAAGCGUAUUAUUUG-- -----------.(((((((.........((((-----------.........))))....((((.((((.......))))..)))))))))))....................-- ( -16.90, z-score = -0.64, R) >droVir3.scaffold_12875 7788452 96 + 20611582 -----------CCACUGC-GCCCCGCUUCCUCG-------GCGCUCGGCGCUGCUCUCUCGUGUCGCUGCACUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGCA -----------.((((((-(((..........)-------))))...(((((...((.((((((...(((((......))))).)))))).)).......)))))....)))).. ( -30.30, z-score = -1.61, R) >droMoj3.scaffold_6496 7347761 102 - 26866924 ------AACCACUCCCCCUCCAGCCGCCCCCUG-------CCAUUCGGCGCUGCUCUCUCGUGUCGCUGCACUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGCA ------...((((.........((((.......-------.....))))(((...((.((((((...(((((......))))).)))))).)).......)))......)))).. ( -22.90, z-score = -1.41, R) >droGri2.scaffold_15245 1297981 102 + 18325388 ------CACUGCUGCUGCCGCUGCUGUUGCCGC-------CCGCUCGACGCAGCUCUCUCGUGUCGCUGCACUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUA ------((((..(((.((.(((((.((((..(.-------...).))))))))).((.((((((...(((((......))))).)))))).))........)))))...)))).. ( -29.20, z-score = -0.03, R) >consensus ____CCUUUCGCUCGCCACCCCGCCGUCCCGC________AUCCUCAUCGUUCGGCUGUCAUGUCGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUG__ .......................................................((.((((((...(((((......))))).)))))).))...................... (-10.94 = -10.76 + -0.18)


| Location | 13,111,759 – 13,111,856 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.08 |
| Shannon entropy | 0.34044 |
| G+C content | 0.45183 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.73 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
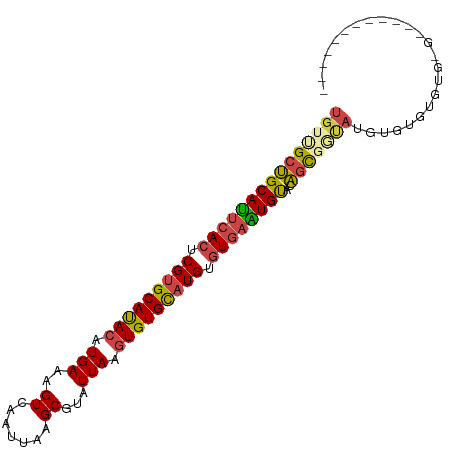
>dm3.chr2R 13111759 97 + 21146708 -CGCAUCCUCAUCGUUCGGCUGUCAUGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCG----------- -.(((.((.........)).)))...((((.(((((((((.(((((((((.(((..((.......))...))).))))))))).))))))))))))).----------- ( -32.70, z-score = -2.55, R) >droEre2.scaffold_4845 9847298 108 - 22589142 -CGCAUCCUCAUCGUUCGGCUGUCAUGUCGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCGGCAUGUGUGUG -.................((...(((((((((((((((((.(((((((((.(((..((.......))...))).))))))))).)))))))..))))))))))...)). ( -41.20, z-score = -3.26, R) >droYak2.chr2R 2780211 106 - 21139217 -CGGGUCCUCAUCGUUCGGCUGUCAUGUCGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCGAUAUAUGUG-- -..((((..........))))...((((((((((((((((.(((((((((.(((..((.......))...))).))))))))).)))))))..))))))))).....-- ( -36.90, z-score = -2.81, R) >droSec1.super_1 10615273 97 + 14215200 -CGGAUCCUCAUCGUUCGGCUGUCAUGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGUG----------- -(((((.......)))))(((((.........((((((((.(((((((((.(((..((.......))...))).))))))))).))))))))))))).----------- ( -30.50, z-score = -2.03, R) >droSim1.chr2R 11844026 97 + 19596830 -CGGAUCCUCAUCGUUCGGCUGUCAUGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCG----------- -(((((.......)))))........((((.(((((((((.(((((((((.(((..((.......))...))).))))))))).))))))))))))).----------- ( -33.10, z-score = -2.66, R) >droAna3.scaffold_13266 6377701 97 - 19884421 CCCAAUCUUCA-CUUCGCGCUGUCAUGUCGCUGCACUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCCUGUGUGAGUGCACAUUC----------- ...........-....((((......).)))(((((((((.((.((((((.(((..((.......))...))).)))))).)).))))))))).....----------- ( -29.00, z-score = -2.39, R) >droWil1.scaffold_181009 2870723 92 - 3585778 ------CUUAACUUUUGGGCCAAAGUGUAGCGGCAUUCACUCGUGCACACAUGAAAGUCAAUUAAGCGUAUUAUUUGUGUAUGUAUAUGUGCAUCGCG----------- ------....(((((......)))))...(((((((.....((((((((.((((..((.......))...)))).)))))))).....))))..))).----------- ( -20.60, z-score = 0.24, R) >consensus _CGGAUCCUCAUCGUUCGGCUGUCAUGUCGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCG___________ ..................(((((.........((((((((.(((((((((.(((..((.......))...))).))))))))).)))))))))))))............ (-26.80 = -26.73 + -0.08)


| Location | 13,111,783 – 13,111,873 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Shannon entropy | 0.38772 |
| G+C content | 0.42248 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.87 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13111783 90 + 21146708 UGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCGGUAUGUGUGUGUGUGAG---------- .((((.(((((((((.(((((((((.(((..((.......))...))).))))))))).)))))))))))))..................---------- ( -31.00, z-score = -2.16, R) >droYak2.chr2R 2780235 100 - 21139217 UGUCGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCGAUAUAUGUGAGUGUGUGUGUGUGGGAG (((((((((((((((.(((((((((.(((..((.......))...))).))))))))).)))))))..))))))))........................ ( -34.30, z-score = -2.35, R) >droSec1.super_1 10615297 88 + 14215200 UGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGUGGUAUGUGUGUAUGAG------------ ((..(((((((((((.(((((((((.(((..((.......))...))).))))))))).)))))))..))))..))............------------ ( -28.40, z-score = -2.06, R) >droSim1.chr2R 11844050 88 + 19596830 UGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCGGUAUGUGUGUGUGAG------------ .((((.(((((((((.(((((((((.(((..((.......))...))).))))))))).)))))))))))))................------------ ( -31.00, z-score = -2.48, R) >droAna3.scaffold_13266 6377725 86 - 19884421 UGUCGCUGCACUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCCUGUGUGAGUGCACAUUCCCAUUUGAGGGUA-------------- ....(.(((((((((.((.((((((.(((..((.......))...))).)))))).)).))))))))))...(((......)))..-------------- ( -29.20, z-score = -2.51, R) >droWil1.scaffold_181009 2870742 73 - 3585778 UGUAGCGGCAUUCACUCGUGCACACAUGAAAGUCAAUUAAGCGUAUUAUUUGUGUAUGUAUAUGUGCAUCGCG--------------------------- ....(((((((.....((((((((.((((..((.......))...)))).)))))))).....))))..))).--------------------------- ( -19.30, z-score = -0.60, R) >consensus UGUUGCUGCAUUCACUCGUGCAUACAUGAAAGUCAAUUAAGCGUAUUAAGUGUGCAUGUGUGAAUGUACAGCGGUAUGUGUGUGUG_G____________ (((((((((((((((.(((((((((.(((..((.......))...))).))))))))).))))))))..)))))))........................ (-25.50 = -25.87 + 0.37)
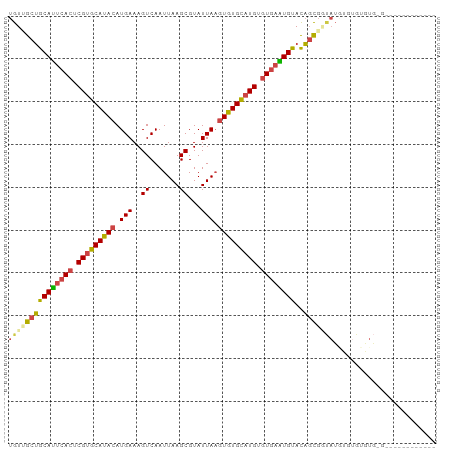
| Location | 13,111,783 – 13,111,873 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Shannon entropy | 0.38772 |
| G+C content | 0.42248 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -13.58 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13111783 90 - 21146708 ----------CUCACACACACACAUACCGCUGUACAUUCACACAUGCACACUUAAUACGCUUAAUUGACUUUCAUGUAUGCACGAGUGAAUGCAGCAACA ----------..................((((..(((((((.(.((((.((........(......)........)).)))).).))))))))))).... ( -19.59, z-score = -2.33, R) >droYak2.chr2R 2780235 100 + 21139217 CUCCCACACACACACACUCACAUAUAUCGCUGUACAUUCACACAUGCACACUUAAUACGCUUAAUUGACUUUCAUGUAUGCACGAGUGAAUGCAGCGACA ..........................((((((..(((((((.(.((((.((........(......)........)).)))).).))))))))))))).. ( -22.69, z-score = -3.25, R) >droSec1.super_1 10615297 88 - 14215200 ------------CUCAUACACACAUACCACUGUACAUUCACACAUGCACACUUAAUACGCUUAAUUGACUUUCAUGUAUGCACGAGUGAAUGCAGCAACA ------------.................(((..(((((((.(.((((.((........(......)........)).)))).).))))))))))..... ( -15.09, z-score = -0.98, R) >droSim1.chr2R 11844050 88 - 19596830 ------------CUCACACACACAUACCGCUGUACAUUCACACAUGCACACUUAAUACGCUUAAUUGACUUUCAUGUAUGCACGAGUGAAUGCAGCAACA ------------................((((..(((((((.(.((((.((........(......)........)).)))).).))))))))))).... ( -19.59, z-score = -2.38, R) >droAna3.scaffold_13266 6377725 86 + 19884421 --------------UACCCUCAAAUGGGAAUGUGCACUCACACAGGCACACUUAAUACGCUUAAUUGACUUUCAUGUAUGCACGAGUGAGUGCAGCGACA --------------..(((......)))....(((((((((.(..(((.((........(......)........)).)))..).)))))))))...... ( -24.29, z-score = -1.98, R) >droWil1.scaffold_181009 2870742 73 + 3585778 ---------------------------CGCGAUGCACAUAUACAUACACAAAUAAUACGCUUAAUUGACUUUCAUGUGUGCACGAGUGAAUGCCGCUACA ---------------------------..((.(((((((((.......(((.(((.....))).)))......)))))))))))((((.....))))... ( -14.22, z-score = -0.20, R) >consensus ____________C_CACACACACAUACCGCUGUACAUUCACACAUGCACACUUAAUACGCUUAAUUGACUUUCAUGUAUGCACGAGUGAAUGCAGCAACA ............................((((..(((((((.(.((((.((........(......)........)).)))).).))))))))))).... (-13.58 = -14.31 + 0.72)

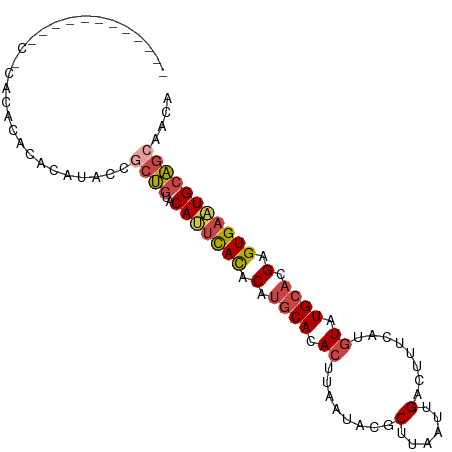

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:22 2011