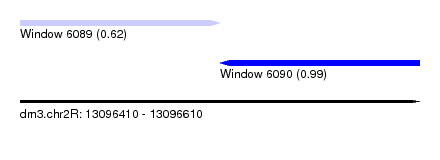
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,096,410 – 13,096,610 |
| Length | 200 |
| Max. P | 0.992886 |

| Location | 13,096,410 – 13,096,510 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.68 |
| Shannon entropy | 0.21717 |
| G+C content | 0.46964 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -12.00 |
| Energy contribution | -11.89 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
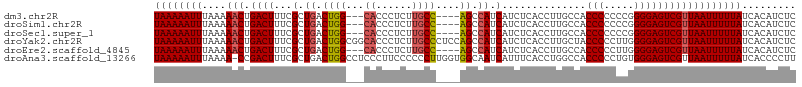
>dm3.chr2R 13096410 100 + 21146708 UAAAAAUUUAAAAACUGACUUUCGCUGACUGG---CACCCUCUUGCC----AGCCAUCAUCUCACCUUGCCACCCCCCCGGGGAGUCGUUAAUUUUUAUCACAUCUC ((((((((....(((.((((...(.((.((((---((......))))----)).)).)..............((((...)))))))))))))))))))......... ( -22.40, z-score = -2.95, R) >droSim1.chr2R 11827282 100 + 19596830 UAAAAAUUUAAAAACUGACUUUCGCUGACUGG---CACCCUCUUGCC----AGCCAUCAUCUCACCUUGCCACCCCCCCGGGGAGUCGUUAAUUUUUAUCACAUCUC ((((((((....(((.((((...(.((.((((---((......))))----)).)).)..............((((...)))))))))))))))))))......... ( -22.40, z-score = -2.95, R) >droSec1.super_1 10599918 100 + 14215200 UAAAAAUUUAAAAACUGACUUUCGCUGACUGG---CACCCUCUUGCC----AGCCAUCAUCUCACCUUGCCACCCCCCCGGGGAGUCGUUAAUUUUUAUCACAUCUC ((((((((....(((.((((...(.((.((((---((......))))----)).)).)..............((((...)))))))))))))))))))......... ( -22.40, z-score = -2.95, R) >droYak2.chr2R 2764925 107 - 21139217 UAAAAAUUUAAAAACUGACUUUCGCUGACUGGCGGCACCCUCUUGCCCUCCAGCCAUCAUCUCACCUUGCUACCCCCUUGGGGAGUCGUUAAUUUUUAUCACAUCUC ........((((((.((((....(.((.((((.((((......))))..)))).)).)..........(((.((((...))))))).)))).))))))......... ( -23.60, z-score = -2.25, R) >droEre2.scaffold_4845 9832027 100 - 22589142 UAAAAAUUUAAAAACUGACUUUCGCUGACUGG---CACCCUCUUGCC----AGCCAUCAUCUCACCUUGCCACCCCCUUGGGGAGUCGUUAAUUUUUAUCACAUCUC ((((((((....(((.((((...(.((.((((---((......))))----)).)).)..............((((...)))))))))))))))))))......... ( -22.40, z-score = -2.87, R) >droAna3.scaffold_13266 6364400 106 - 19884421 UAAAAAUUUAAAA-CCGACUUUCGCUGACUGGCCUCCCUUCCCCCCUUGGUGGCAAUCAUUUCACCUGGCCACCCCCUGUGGGAGUCGUUAAUUUUUAUCACCCCUU ((((((((.....-.(((((..(((.(..(((((..............(((((........))))).)))))..)...)))..)))))..))))))))......... ( -26.96, z-score = -2.50, R) >consensus UAAAAAUUUAAAAACUGACUUUCGCUGACUGG___CACCCUCUUGCC____AGCCAUCAUCUCACCUUGCCACCCCCCCGGGGAGUCGUUAAUUUUUAUCACAUCUC ((((((((....(((.((((...(((....((.............))....)))..................(((.....))))))))))))))))))......... (-12.00 = -11.89 + -0.11)
| Location | 13,096,510 – 13,096,610 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Shannon entropy | 0.38750 |
| G+C content | 0.47592 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -21.91 |
| Energy contribution | -23.58 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
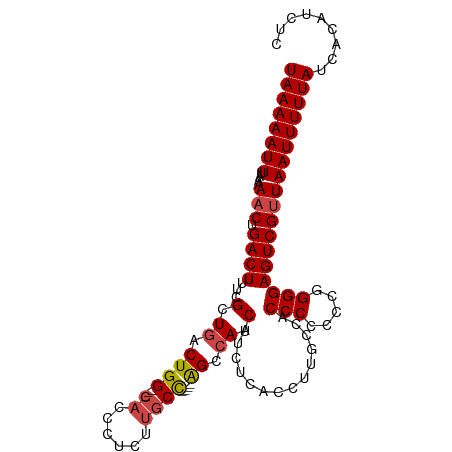
>dm3.chr2R 13096510 100 - 21146708 AAAAUGGGCAGACACCUGCUCCACUGGUA------CUGGUGCUCAAUGCAUUUUU--UGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCGGUAUCAAG ....(((((((....)))).))).(((((------(((((((((..((((((((.--.....))))))))(((..............)))..)))))))))))))).. ( -40.44, z-score = -4.88, R) >droSim1.chr2R 11827382 100 - 19596830 AAAAUGGGCAGACACCUGCUCCACUGGAA------CUGGUGCUCAAUGCAUUUUU--GGAGUGUAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAGUAUCAAG ....(((((((....)))).))).(((.(------(((((((((..((((((...--.......))))))(((..............)))..)))))))))).))).. ( -33.84, z-score = -3.07, R) >droSec1.super_1 10600018 100 - 14215200 AAAAUGGGCAGACACCUGCUCCAAUGGAA------CUGGUGCUCAAUGCAUUUUU--GGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAGUAUCAGG ....(((((((....)))).))).(((.(------(((((((((..((((((((.--.....))))))))(((..............)))..)))))))))).))).. ( -37.24, z-score = -4.02, R) >droYak2.chr2R 2765032 94 + 21139217 GAAAUGGGCAGACACCUGCUCC------A------CUGGUGCUCAAUGCAUUUUU--AGAGUGGAAUGCAGUCAAAAACAGAGGACCAACAAGACUACCAGUAUCAAG ((...((((((....)))).))------(------((((((.((..((((((((.--.....))))))))(((..........)))......)).))))))).))... ( -27.40, z-score = -2.14, R) >droEre2.scaffold_4845 9832127 94 + 22589142 GAAAUGGGCAGACACCUGCUCC------A------CUGGUGCUCAAUGCAUUUUU--GGAGUGGAAUGCAGUCUAAAACAGAGGACCAACCAGAGUACAAGUAUCAAG ((...((((((....)))).))------(------((.((((((..((((((((.--.....))))))))((((........))))......)))))).))).))... ( -29.30, z-score = -2.07, R) >dp4.chr3 16679381 100 - 19779522 GAAAUGGCCAAACACCUGCCCCAUGGCUACCAUGGCUGGCCCCCAAUGCAGUUCUCAGCAUCAGAAUGCAGCCAAAAACAGAGACAGAGACAGGAGAGAA-------- ....((((.........(((((((((...))))))..)))......((((.((((.......))))))))))))..........................-------- ( -21.80, z-score = 0.72, R) >consensus AAAAUGGGCAGACACCUGCUCCA_UGG_A______CUGGUGCUCAAUGCAUUUUU__GGAGUGGAAUGCAGUCAAAAACAGAGGACCAACAAGAGUACCAGUAUCAAG .....((((((....))))))..............(((((((((..((((((((........))))))))(((..........)))......)))))))))....... (-21.91 = -23.58 + 1.67)

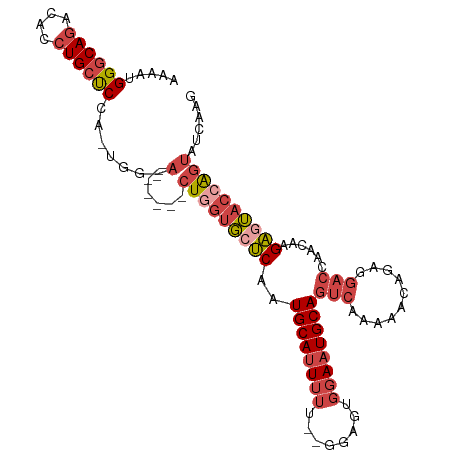
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:19 2011