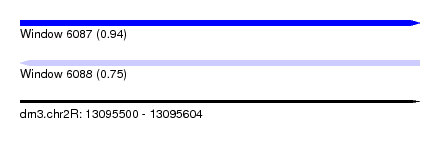
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,095,500 – 13,095,604 |
| Length | 104 |
| Max. P | 0.939536 |

| Location | 13,095,500 – 13,095,604 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 86.05 |
| Shannon entropy | 0.26302 |
| G+C content | 0.41663 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -28.75 |
| Energy contribution | -30.03 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939536 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
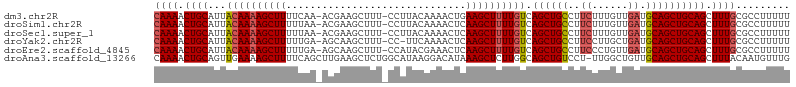
>dm3.chr2R 13095500 104 + 21146708 AAAAAGGCGCAAAGCUGCAGCUGCAUCAACAAAGAAGGCAGCUGACAAAAGCUUCAGUUUUGUAAGG-AAAGCUUCGU-UUGAAAAGCUUUUGUAAUGCAGUUUUG .........(((((((((((((((.((......))..)))))).(((((((((((((...((.(((.-....))))).-))))..)))))))))...))))))))) ( -34.20, z-score = -1.88, R) >droSim1.chr2R 11826382 104 + 19596830 AAAAAGGCGCAAAGCUGCAGCUGCAUCAACAAAGAAGGCAGCUGACAAAAGCUUGAGUUUUGUAAGG-AAAGCUUCGU-UUAAAAAGCUUUUGUAAUGCAGUUUUG .........(((((((((((((((.((......))..)))))).(((((((((((((((((......-)))))))...-.....))))))))))...))))))))) ( -35.60, z-score = -2.69, R) >droSec1.super_1 10599014 104 + 14215200 AAAAAGGCGCAAAGCUGCAGCUGCAUCAACAAAGAAGGCAGCUGACAAAAGCUUGAGUUUUGUAAGG-AAAGCUUCGU-UUAAAAAGCUUUUGUAAUGCAGUUUUG .........(((((((((((((((.((......))..)))))).(((((((((((((((((......-)))))))...-.....))))))))))...))))))))) ( -35.60, z-score = -2.69, R) >droYak2.chr2R 2764031 103 - 21139217 AAAAAGGCGCAAAGCUGCAGCUGCAUCAGCAAGGAAGGCAGCUGACAAAAGCUUGAGUUUUGAA-GG-AAAGCUUGCU-UCAAAAAGCUUUUGUAAUGCAGUUUUG .........(((((((((((((((.((......))..)))))).((((((((((....((((((-(.-(.....).))-)))))))))))))))...))))))))) ( -38.00, z-score = -2.14, R) >droEre2.scaffold_4845 9831144 104 - 22589142 AAAAAGGCGCAAAGCUGCAGCUGCAUCAACAGGGAAGGCAGCUGACAAAAGCUUGAGUUUCGUAUGG-AAAGCUUGCU-UCAAAAAGCUUUUGUAAUGCAGUUUUG .........(((((((((((((((.((......))..)))))).((((((((((((((((.......-.))))))...-.....))))))))))...))))))))) ( -33.90, z-score = -1.32, R) >droAna3.scaffold_13266 6363757 105 - 19884421 CAAACAUUGUAAAGCUGCAGCUGCAACAGCCAA-AGGACAGCUGCCAAGAGCUUUAUGUCCUUAUGCCAGAGCUUCAAGCUGAAAAGCUUUUCAACUGCAGUUUUG .........(((((((((((..((....((..(-((((((...((.....))....)))))))..))....))...(((((....))))).....))))))))))) ( -32.60, z-score = -1.27, R) >consensus AAAAAGGCGCAAAGCUGCAGCUGCAUCAACAAAGAAGGCAGCUGACAAAAGCUUGAGUUUUGUAAGG_AAAGCUUCGU_UUAAAAAGCUUUUGUAAUGCAGUUUUG .........(((((((((((((((.((......))..)))))).(((((((((((((((((.......))))))).........))))))))))...))))))))) (-28.75 = -30.03 + 1.28)

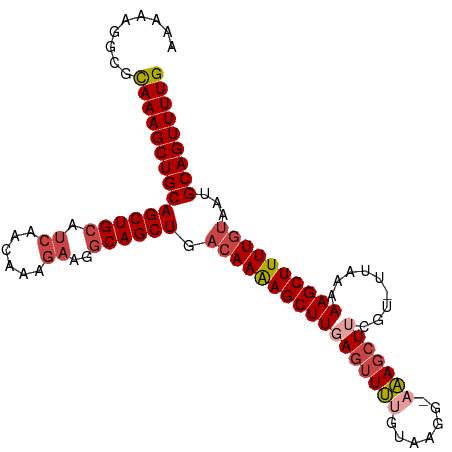
| Location | 13,095,500 – 13,095,604 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.05 |
| Shannon entropy | 0.26302 |
| G+C content | 0.41663 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -22.49 |
| Energy contribution | -22.91 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745925 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 13095500 104 - 21146708 CAAAACUGCAUUACAAAAGCUUUUCAA-ACGAAGCUUU-CCUUACAAAACUGAAGCUUUUGUCAGCUGCCUUCUUUGUUGAUGCAGCUGCAGCUUUGCGCCUUUUU ((((.((((...(((((((((((....-...(((....-.)))........))))))))))).((((((..((......)).)))))))))).))))......... ( -30.06, z-score = -2.39, R) >droSim1.chr2R 11826382 104 - 19596830 CAAAACUGCAUUACAAAAGCUUUUUAA-ACGAAGCUUU-CCUUACAAAACUCAAGCUUUUGUCAGCUGCCUUCUUUGUUGAUGCAGCUGCAGCUUUGCGCCUUUUU ((((.((((...((((((((((.....-...(((....-.))).........)))))))))).((((((..((......)).)))))))))).))))......... ( -27.93, z-score = -2.17, R) >droSec1.super_1 10599014 104 - 14215200 CAAAACUGCAUUACAAAAGCUUUUUAA-ACGAAGCUUU-CCUUACAAAACUCAAGCUUUUGUCAGCUGCCUUCUUUGUUGAUGCAGCUGCAGCUUUGCGCCUUUUU ((((.((((...((((((((((.....-...(((....-.))).........)))))))))).((((((..((......)).)))))))))).))))......... ( -27.93, z-score = -2.17, R) >droYak2.chr2R 2764031 103 + 21139217 CAAAACUGCAUUACAAAAGCUUUUUGA-AGCAAGCUUU-CC-UUCAAAACUCAAGCUUUUGUCAGCUGCCUUCCUUGCUGAUGCAGCUGCAGCUUUGCGCCUUUUU ((((.((((...(((((((((((((((-((........-.)-))))))....)))))))))).((((((..((......)).)))))))))).))))......... ( -31.50, z-score = -2.18, R) >droEre2.scaffold_4845 9831144 104 + 22589142 CAAAACUGCAUUACAAAAGCUUUUUGA-AGCAAGCUUU-CCAUACGAAACUCAAGCUUUUGUCAGCUGCCUUCCCUGUUGAUGCAGCUGCAGCUUUGCGCCUUUUU ((((.((((...((((((((((...((-((....))))-......(.....))))))))))).((((((..((......)).)))))))))).))))......... ( -27.20, z-score = -1.17, R) >droAna3.scaffold_13266 6363757 105 + 19884421 CAAAACUGCAGUUGAAAAGCUUUUCAGCUUGAAGCUCUGGCAUAAGGACAUAAAGCUCUUGGCAGCUGUCCU-UUGGCUGUUGCAGCUGCAGCUUUACAAUGUUUG .(((.(((((((((...((((((.......))))))..(((..(((((((....((.....))...))))))-)..)))....))))))))).))).......... ( -32.60, z-score = -0.26, R) >consensus CAAAACUGCAUUACAAAAGCUUUUUAA_ACGAAGCUUU_CCUUACAAAACUCAAGCUUUUGUCAGCUGCCUUCUUUGUUGAUGCAGCUGCAGCUUUGCGCCUUUUU ((((.((((...((((((((((..............................)))))))))).((((((..((......)).)))))))))).))))......... (-22.49 = -22.91 + 0.42)
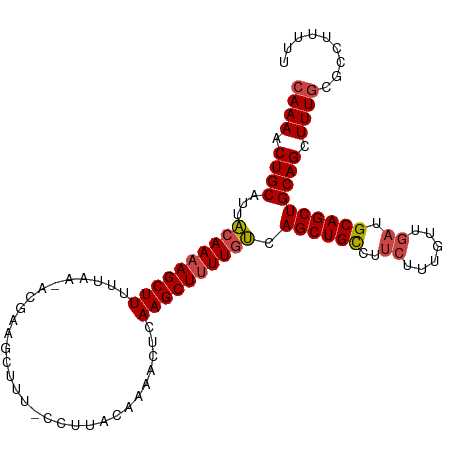
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:17 2011