
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,086,462 – 13,086,641 |
| Length | 179 |
| Max. P | 0.624524 |

| Location | 13,086,462 – 13,086,572 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.78 |
| Shannon entropy | 0.66514 |
| G+C content | 0.48632 |
| Mean single sequence MFE | -30.59 |
| Consensus MFE | -10.35 |
| Energy contribution | -9.59 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
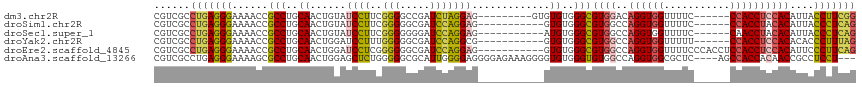
>dm3.chr2R 13086462 110 - 21146708 CAGGUGGUUUUC------CCACCUCCACAUUACCUUCGGCCCCGU--GGUGCGGUUGCCGUCGGGCUGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG- ..(((((.....------)))))(((((......((((((((..(--(((......))))..)))))))).......(((.(((((((....))))))).))))))))..........- ( -43.20, z-score = -3.48, R) >droSim1.chr2R 11817514 110 - 19596830 CAGGUGGUUUUC------CCACCUACACAUUACCCUCAGCCCCGU--GGUGCGGUUGCCGUCGGGCUGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG- .((((((.....------)))))).(((.......(((((((..(--(((......))))..)))))))........(((.(((((((....))))))).))))))............- ( -39.90, z-score = -2.84, R) >droSec1.super_1 10590023 110 - 14215200 CAGGUGGUUUUC------CAACCUACACAUUACCCUCAGCCCCGU--GGUGCGGUUGCCGUCGUGCUGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG- .......(((((------((.(((..............((.(((.--....)))..)).((((((.((((((((((((....))))).))))))))))))))))))))))).......- ( -31.10, z-score = -0.88, R) >droYak2.chr2R 2750662 112 + 21139217 CAGGUGGUUUUU------CCACCUCCACACACCCUUUAGCUCUGUGCGGUGCGGUUGCCGUCGGGCUGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG- ..(((((.....------)))))(((((......((((((.(((.(((((......)))))))))))))).......(((.(((((((....))))))).))))))))..........- ( -40.60, z-score = -2.90, R) >droEre2.scaffold_4845 9822109 116 + 22589142 CAGGUGGUUUUCCCACCUCCACCUCCACAUUCCCUUCAGCCCUGU--GGUGCGGUUGCCGUCGGGCUGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG- .((((((.....)))))).....(((((......((((((((..(--(((......))))..)))))))).......(((.(((((((....))))))).))))))))..........- ( -44.60, z-score = -3.85, R) >droAna3.scaffold_13266 6354828 109 + 19884421 CAGGUGGCGCU-------CAGCCACCACAACCGCCUCCUCCCAGC--GGUGCGGUUGCCGUCAGGCUAAAAUUUAUGCUUCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAGG- ..((((((...-------..)))))).(((((((..((.......--)).)))))))(((((((((..........))))..((((((....))))))))).))..............- ( -36.60, z-score = -1.35, R) >dp4.chr3 16670818 84 - 19779522 ---------------------------------CGACACACCCGCACAGUGCGGUUGCCGUC-GGUGGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG- ---------------------------------........((((.....))))(..(((((-(((((((.(((((((....))))))).)))))).)))).))..)...........- ( -26.40, z-score = -1.72, R) >droPer1.super_4 7061840 84 + 7162766 ---------------------------------CGACACACCCGCACAGUGCGGUUGCCGUC-GGUGGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG- ---------------------------------........((((.....))))(..(((((-(((((((.(((((((....))))))).)))))).)))).))..)...........- ( -26.40, z-score = -1.72, R) >droWil1.scaffold_181009 2815987 86 + 3585778 --------------------------------CAUGAGAACUUAUUGUGGAAAAUUGGCUGUUGGCUGAAAUUUAUGUUGCAGUGUGAAAUUUCAUGCGACAGGUGAAAAGGCAAAAU- --------------------------------((..(.......)..)).....(((.((.((.(((........(((((((.((........)))))))))))).)).)).)))...- ( -15.20, z-score = 0.24, R) >droMoj3.scaffold_6496 7313798 84 + 26866924 ---------------------------------CAGCUGGUUUACCCCAUGCGGUUGCCGUC-GCCCAAAAUUUAUGUUGCAGCAUGAAAUUUACCGCGGCAGGUGGAAAAGAGGCAG- ---------------------------------..(((..((((((((....)).(((((.(-(...(((.(((((((....))))))).)))..))))))))))))).....)))..- ( -24.00, z-score = 0.00, R) >droGri2.scaffold_15245 1267824 86 - 18325388 --------------------------------ACAUUUGGUAUACCCCAUGCGGUUGCCGUC-GGGCAAAAUUUAUGUUUCAGUGCGAAAUUUUACGCGACAGGUGGAAAAAGGGCGAC --------------------------------.(((((((((.(((......)))))))(((-(.(.(((((((.(((......)))))))))).).)))))))))............. ( -20.90, z-score = 0.36, R) >droVir3.scaffold_12875 7754923 84 - 20611582 ---------------------------------CAGGUGGCUUACCCCAUGCGGUUGCCGUC-GGCCAAAAUUUAUGUUGCAGUAUGAAAUUUAAAGCGACAGGUGAAAAAAGAGCAG- ---------------------------------..((.((....)))).((((((((....)-))))........((((((..((.......))..))))))............))).- ( -18.20, z-score = 0.40, R) >consensus ________________________________CCUUCAGCCCUGC_CGGUGCGGUUGCCGUC_GGCUGAAAUUUAUGCUGCUGUGUGAAAUUUUACGCGACAGGUGGAAAAAUAAAAG_ .........................................((((....((((...(((....))).(((((((((((....))))).)))))).))))....))))............ (-10.35 = -9.59 + -0.76)


| Location | 13,086,539 – 13,086,641 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Shannon entropy | 0.34954 |
| G+C content | 0.62205 |
| Mean single sequence MFE | -44.68 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.83 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
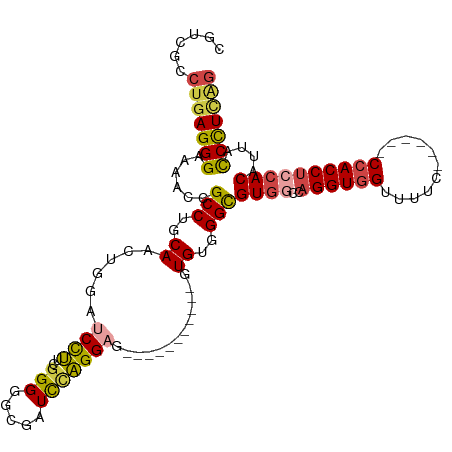
>dm3.chr2R 13086539 102 - 21146708 CGUCGCCUGAGGGAAAACCGCCUGCAACUGUAUCCUUCGGGGCCGAUCUAGGAG---------GUGUGUGGGCGUGGACAGGUGGUUUUC------CCACCUCCACAUUACCUUCGG ......(((((((......(((..(((((...(((....)))((......)).)---------)).))..)))(((((..(((((.....------))))))))))....))))))) ( -42.40, z-score = -1.69, R) >droSim1.chr2R 11817591 100 - 19596830 CGUCGCCUGAGGGAAAACCGCCUGCAACUGUAUCCUUCGGGGGCGAUCCAGGAG-----------GUGUGGGCGUGGCCAGGUGGUUUUC------CCACCUACACAUUACCCUCAG ......(((((((......(((..((.......(((((.(((....))).))))-----------)))..)))(((...((((((.....------)))))).)))....))))))) ( -43.01, z-score = -1.87, R) >droSec1.super_1 10590100 100 - 14215200 CGUCGCCUGAGGGAAAACCGCCUGCAACUGUAUCCUUCGGGGGGGAUCCAGGAG-----------AUGUGGGCGUGGCCAGGUGGUUUUC------CAACCUACACAUUACCCUCAG ......(((((((......(((..((.(((.((((((....)))))).)))...-----------.))..)))(((...((((((....)------).)))).)))....))))))) ( -42.60, z-score = -2.02, R) >droYak2.chr2R 2750741 100 + 21139217 CGUCGCCUGAGGGAAAACCGCCUGCAACUGGAUCCUUUGGGGGCGAUCCAGGCG-----------GUGUGGGCGUGGCCAGGUGGUUUUU------CCACCUCCACACACCCUUUAG ......(((((((......(((..((.(((((((((.....)).)))))))...-----------.))..)))((((..((((((.....------))))))))))....))))))) ( -46.10, z-score = -2.25, R) >droEre2.scaffold_4845 9822186 106 + 22589142 CGUCGCCUGAGGGAAAACCGCCUGCAACUGGAUCCUCGGGGGGCGAUCCAGGAG-----------GUGUGGGCGUGGCCAGGUGGUUUUCCCACCUCCACCUCCACAUUCCCUUCAG ((((.((((((((....(((........))).)))))))).))))....(((..-----------(((((((.((((..((((((.....)))))))))).)))))))..))).... ( -51.10, z-score = -2.38, R) >droAna3.scaffold_13266 6354905 110 + 19884421 CGUCGCCUGAGGGAAAAGCGCCUGCAACUGGAGCUCUGGGGGCGCAUUGGGGAGGGGAGAAAGGGGUGUGGGUGUGGCCAGGUGGCGCUC----AGCCACCACAACCGCCUCCU--- (.((.((..(.......((((((.((..........)).)))))).)..)))).)......(((((.((((.(((.....((((((....----.))))))))).)))))))))--- ( -42.90, z-score = 1.48, R) >consensus CGUCGCCUGAGGGAAAACCGCCUGCAACUGGAUCCUUCGGGGGCGAUCCAGGAG___________GUGUGGGCGUGGCCAGGUGGUUUUC______CCACCUCCACAUUACCCUCAG ......(((((((......(((..((......((((..(((.....))))))).............))..)))((((..((((((...........))))))))))....))))))) (-30.40 = -30.83 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:15 2011