
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,083,656 – 13,083,763 |
| Length | 107 |
| Max. P | 0.843263 |

| Location | 13,083,656 – 13,083,763 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.72 |
| Shannon entropy | 0.50759 |
| G+C content | 0.48038 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -12.40 |
| Energy contribution | -14.15 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
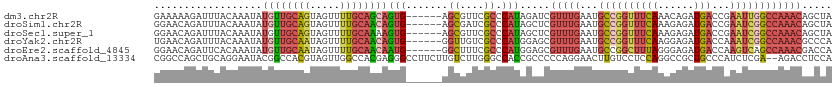
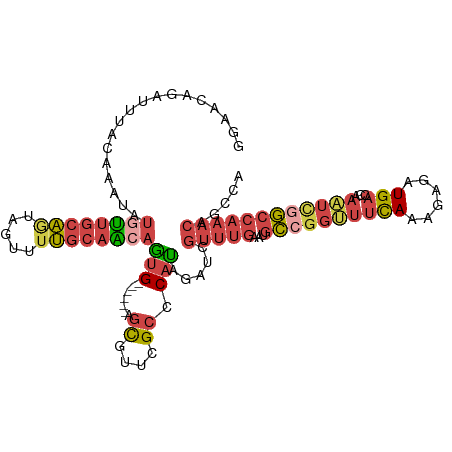
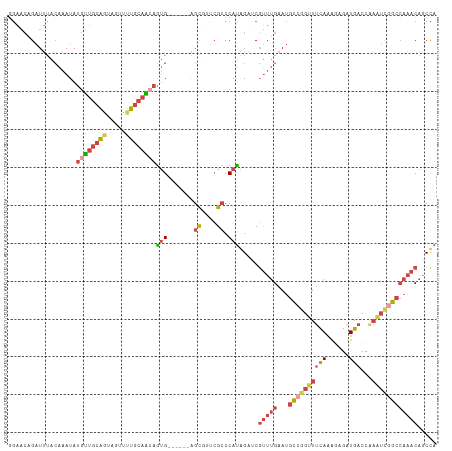
>dm3.chr2R 13083656 107 - 21146708 GAAAAAGAUUUACAAAUAUGUUGCAGUAGUUUUGCAGCAGUG------AGCGUUCGCCCAUAGAUCGUUUGAAUGCCGGUUUCAAACAGAUGACCGAAUUGGCCAAACAGCUA ......(((((.......((((((((.....))))))))(((------.((....)).))))))))(((((...((((((((((......)))...))))))))))))..... ( -27.10, z-score = -1.57, R) >droSim1.chr2R 11814652 107 - 19596830 GGAACAGAUUUACAAAUAUGUUGCAGUAGUUUUGCAACAGUG------AGCGAUCGCCCAUAGCUCGUUUGAAUGCCGGUUUCAAAGAGAUGACCGAAUCGGCCAAACAGCUA ((................((((((((.....))))))))(((------(....)))))).(((((.(((((...((((((((((......)))...))))))))))))))))) ( -32.10, z-score = -2.68, R) >droSec1.super_1 10587150 107 - 14215200 GGAACAGAUUUACAAAUAUGUUGCAGUAGUUUUGCAAAAGUG------AGCGUUCGCCCAUAGCUCGUUUGAAUGCCGGUUUCAAAGAGAUGACCGAAUCGGCCAAACAGCUA .((((.(.(((((.......((((((.....))))))..)))------))))))).....(((((.(((((...((((((((((......)))...))))))))))))))))) ( -27.70, z-score = -1.25, R) >droYak2.chr2R 2747728 107 + 21139217 UGAACAGAUUUACAAAUAUGUUGCAAUAGUUUUGCAACAGUG------GGUUGUCGCCCAUGGAGCGUUUGAAUGCCGGUUUCAAGGAGAUGACCAAAUCGGCCAAACGCCCA ..................((((((((.....))))))))(((------(((....))))))((.(((((((...((((((((...((......))))))))))))))))))). ( -39.70, z-score = -3.87, R) >droEre2.scaffold_4845 9819246 107 + 22589142 GGAACAGAUUCACAAAUAUGUUGCAAUAGUUUUGCAACAAUG------GGCUUUCGCCCAUGGAGCGUUUGAAUGCCGGCUUUAGGGAGAUGACCAAGUCAGCCAAACGACCA ((......(((.......((((((((.....))))))))(((------(((....))))))))).((((((...((.(((((..((.......))))))).)))))))).)). ( -34.90, z-score = -2.58, R) >droAna3.scaffold_13334 520698 111 + 1562580 CGGCCAGCUGCAGGAAUACGGCCACGUAGUUGGCCACGAGGGCCUUCUUGUCUUGGGCCACCGCCCCCAGGAACUUGUCCUCCAGGCCGCUGCCCAUCUCGA--AGACCUCCA .((((((((((.((.......))..))))))))))..((((..((((..(...(((((..(.(((...((((.....))))...))).)..))))).)..))--)).)))).. ( -47.50, z-score = -2.15, R) >consensus GGAACAGAUUUACAAAUAUGUUGCAGUAGUUUUGCAACAGUG______AGCGUUCGCCCAUAGAUCGUUUGAAUGCCGGUUUCAAAGAGAUGACCAAAUCGGCCAAACAGCCA ..................((((((((.....))))))))...........................(((((...((((((((((......)))...))))))))))))..... (-12.40 = -14.15 + 1.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:13 2011