| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,070,268 – 13,070,364 |
| Length | 96 |
| Max. P | 0.665934 |

| Location | 13,070,268 – 13,070,364 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 64.38 |
| Shannon entropy | 0.72648 |
| G+C content | 0.54416 |
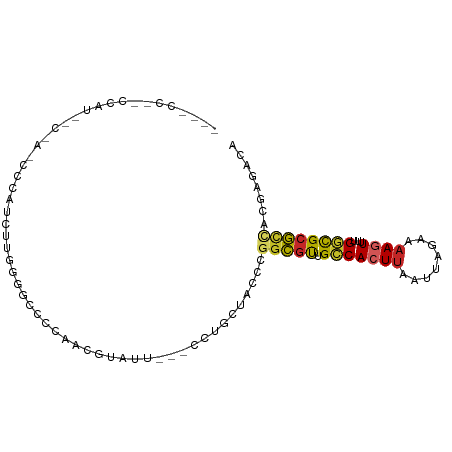
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -9.14 |
| Energy contribution | -9.14 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.50 |
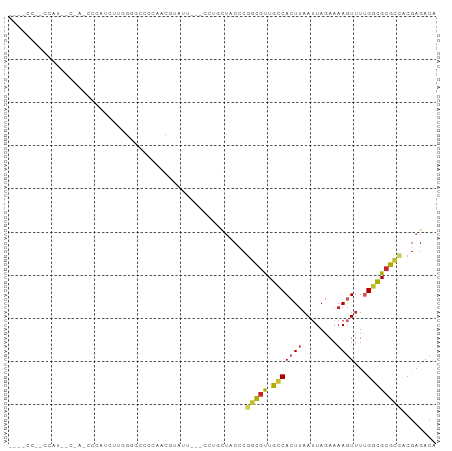
| Mean z-score | -0.84 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.665934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
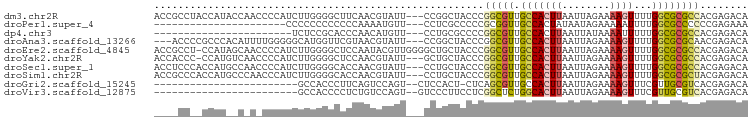
>dm3.chr2R 13070268 96 - 21146708 ACCGCCUACCAUACCAACCCCAUCUUGGGGCUUCAACGUAUU---CCGGCUACCCGGCGUUGCCACUUAAUUAGAAAAGUUUUGGCGCGCCACGAGACA ...(((....((((...((((.....)))).......)))).---..))).....(((((.((((...((((.....)))).)))))))))........ ( -25.40, z-score = -0.73, R) >droPer1.super_4 7050854 74 + 7162766 ----------------------CCCCCCCCCCCCAAAAUGUU---CCUCGCCCCCGCGGUUGCCACUAUAAUAGAAAAAUUUUGGCGCCCCCCGAGAAA ----------------------....................---.(((((....))((.(((((..((.........))..))))).))...)))... ( -11.10, z-score = -1.16, R) >dp4.chr3 16659826 73 - 19779522 -----------------------UCUCCGCACCCAACAUGUU---CCUGCGCCCCGGCGUUGCCACUUAAUUAUAAAAUUUUUGGCGCGCCACGAGACA -----------------------((((((((...........---..))))....(((((.((((...((((....))))..)))))))))..)))).. ( -20.22, z-score = -2.33, R) >droAna3.scaffold_13266 6341190 93 + 19884421 ---ACCCCGCCCACAUUUUGGGGGCAUGGUUCGUAACGUAUU---CCGGCUACCCGGCGUUGCCACUUAAUUAGAAAAGUUUUGGCGCGCAACGAGACA ---(((..((((.(.....).))))..)))..((..(((...---.(((....)))((((.((((...((((.....)))).)))))))).)))..)). ( -26.00, z-score = 0.53, R) >droEre2.scaffold_4845 9806256 98 + 22589142 ACCGCCU-CCAUAGCAACCCCAUCUUGGGGCUCCAAUACGUUGGGGCUGCUACCCGGCGUUGCCACUUAAUUAGAAAAGUUUUGGCGCGCCACGAGACA ...(.((-(..(((((..(((.....)))(((((((....))))))))))))...(((((.((((...((((.....)))).)))))))))..))).). ( -35.90, z-score = -2.07, R) >droYak2.chr2R 2734383 95 + 21139217 ACCACCC-CCAUGUCAACCCCAUCUUGGGGCUCCAACGUAUU---GCUGCUACCCGGCGUUGCCACUUAAUUAGAAAAGUUUUGGCGCGCCACGAGACA .......-...((((..((((.....))))............---..........(((((.((((...((((.....)))).)))))))))....)))) ( -25.20, z-score = -0.75, R) >droSec1.super_1 10574351 96 - 14215200 ACCUCCCACCAUGCCAACCCCAUCUUGGGGCACCAACGUAUU---CCUGCUACCCGGCGUUGCCACUUAAUUAGAAAAGUUUUGGCGCGCCACGAGACA ..(((............((((.....))))............---..........(((((.((((...((((.....)))).)))))))))..)))... ( -23.80, z-score = -0.63, R) >droSim1.chr2R 11798784 96 - 19596830 ACCGCCCACCAUGCCCAACCCAUCUUGGGGCACCAACGUAUU---CCUGCUACCCGGCGUUGCCACUUAAUUAGAAAAGUUUUGGCGCGCUACGAGACA ...((((...(((.......)))....))))...........---..........(((((.((((...((((.....)))).)))))))))........ ( -19.40, z-score = 1.34, R) >droGri2.scaffold_15245 1252838 72 - 18325388 ------------------------GCCACCCUUCAGUCCAGU--CUCCACU-CUCAGCGUUGCCACUUAAUUAGAAAAGUUUCGUUGCGUCACGAGACA ------------------------................((--(((....-(.(((((..((..((.....))....))..))))).)....))))). ( -13.50, z-score = -1.46, R) >droVir3.scaffold_12875 7740126 73 - 20611582 ------------------------GCCACCCCUCUGUCCAGU--GUCCCUUCCUCGGCUCUGGCACUUAAUUAGAAAAGUUUCGUUGCGUCACGAGACA ------------------------...........((((((.--(((........))).)))).))............(((((((......))))))). ( -16.80, z-score = -1.14, R) >consensus ____CC__CCAU__C_A_CCCAUCUUGGGGCCCCAACGUAUU___CCUGCUACCCGGCGUUGCCACUUAAUUAGAAAAGUUUUGGCGCGCCACGAGACA .......................................................(((((.(((((((........))))...))))))))........ ( -9.14 = -9.14 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:11 2011