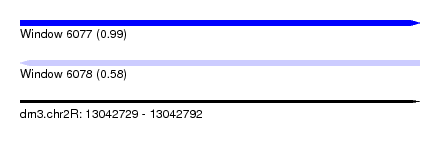
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,042,729 – 13,042,792 |
| Length | 63 |
| Max. P | 0.986562 |

| Location | 13,042,729 – 13,042,792 |
|---|---|
| Length | 63 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 67.07 |
| Shannon entropy | 0.70344 |
| G+C content | 0.33456 |
| Mean single sequence MFE | -10.77 |
| Consensus MFE | -5.97 |
| Energy contribution | -5.72 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
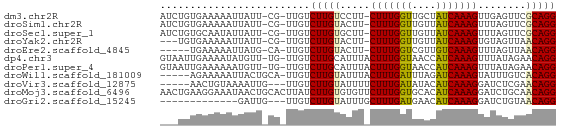
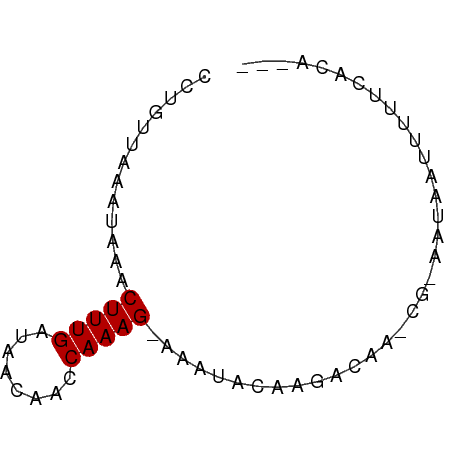
>dm3.chr2R 13042729 63 + 21146708 AUCUGUGAAAAAUUAUU-CG-UUGUCUUGUCCUU-CUUUGGUUGCUAUCAAAGUUGAGUUCGCAGG ..(((((((...(((..-..-(((......((..-....)).......)))...))).))))))). ( -11.92, z-score = -1.84, R) >droSim1.chr2R 11776250 63 + 19596830 AUCUGUGAAAAAUUAUU-CG-UUGUCUUGUACUU-CUUUGGUUGUUAUCAAAGUUUAGUUCGCAGG ..(((((((........-.(-(........))..-(((((((....))))))).....))))))). ( -12.40, z-score = -2.88, R) >droSec1.super_1 10553008 63 + 14215200 AUCUGUGCAAUAUUAUU-CG-UUGUCUUGUGCUU-CUUUGGUUGUUAUCAAAGUUUAGUUCGCAGG ..(((((((((......-.)-))))...(.(((.-(((((((....)))))))...))).))))). ( -12.40, z-score = -2.58, R) >droYak2.chr2R 2711786 60 - 21139217 ---UGUGAAAAAUUAUU-CG-UUGUCUUGUACUU-CUUUGGUUGUUAUCAAAGUGUAGUUAACAGG ---..............-..-....((((((((.-(((((((....)))))))...)))..))))) ( -9.70, z-score = -1.44, R) >droEre2.scaffold_4845 9783059 58 - 22589142 -----UGAAAAAUUAUG-CA-UUGUCUUGUACUU-CUUUGGUCGUUGUCAAAGUUUAGUUAACAGG -----............-..-....((((((((.-((((((......))))))...)))..))))) ( -8.10, z-score = -0.41, R) >dp4.chr3 16639912 64 + 19779522 GUAAUUGAAAAUAUGUU-UG-UUGUCUUGCAUUUACUUUGGUAACCAUCAAAGUUUAUAGAACAGG .............((((-((-(......))((..((((((((....))))))))..)).))))).. ( -8.90, z-score = -0.02, R) >droPer1.super_4 7031616 64 - 7162766 GUAAUUGAAAAAAUGUU-UG-UUGUCUUGCAUUUACUUUGGUAACCAUCAAAGUUUAUAGAACAGG ....(((...((((((.-.(-....)..))))))((((((((....))))))))........))). ( -9.50, z-score = -0.28, R) >droWil1.scaffold_181009 2764202 60 - 3585778 -----AGAAAAAUUACUGCA-UUGUCUUGUAUUUACUUUGAUUUAGAUCAAAGUAUUUGUCACAGG -----...............-....(((((((.(((((((((....)))))))))...)).))))) ( -11.20, z-score = -1.48, R) >droVir3.scaffold_12875 7710806 58 + 20611582 -----AACUGUAAAAUUG---UUGUCUUGUAUUUUCUUUGAUAUACAUCAAAGGAUCUCGAACAGG -----.............---((((.(((....(((((((((....)))))))))...))))))). ( -10.20, z-score = -1.28, R) >droMoj3.scaffold_6496 7264382 66 - 26866924 AACUGAAGGAAAUAACUGCACUUAUCUUGUGUGUUCUUUGGUGCACAUCAAAGGAUCUGCAACAGG ..(((..(((.......((((.......)))).(((((((((....))))))))))))....))). ( -14.80, z-score = -0.89, R) >droGri2.scaffold_15245 1229998 50 + 18325388 -------------GAUUG---UUGUCUUGUAUUUGCUUUGAUGAACAUCAAAGGAUCUGUAACAGG -------------.....---....(((((((.(.(((((((....))))))).)...)).))))) ( -9.40, z-score = -0.72, R) >consensus ___UGUGAAAAAUUAUU_CG_UUGUCUUGUAUUU_CUUUGGUUGACAUCAAAGUUUAUUUAACAGG .........................(((((.....(((((((....)))))))........))))) ( -5.97 = -5.72 + -0.25)
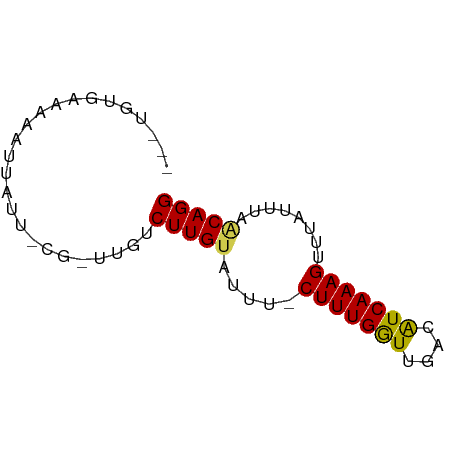
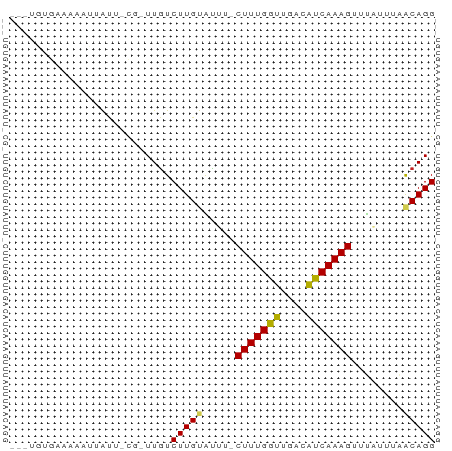
| Location | 13,042,729 – 13,042,792 |
|---|---|
| Length | 63 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 67.07 |
| Shannon entropy | 0.70344 |
| G+C content | 0.33456 |
| Mean single sequence MFE | -6.55 |
| Consensus MFE | -2.91 |
| Energy contribution | -2.91 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13042729 63 - 21146708 CCUGCGAACUCAACUUUGAUAGCAACCAAAG-AAGGACAAGACAA-CG-AAUAAUUUUUCACAGAU .(((.(((.....(((((........)))))-..(.......)..-..-........))).))).. ( -6.80, z-score = -0.97, R) >droSim1.chr2R 11776250 63 - 19596830 CCUGCGAACUAAACUUUGAUAACAACCAAAG-AAGUACAAGACAA-CG-AAUAAUUUUUCACAGAU .(((.(((.....(((((........)))))-..((.....))..-..-........))).))).. ( -7.50, z-score = -2.59, R) >droSec1.super_1 10553008 63 - 14215200 CCUGCGAACUAAACUUUGAUAACAACCAAAG-AAGCACAAGACAA-CG-AAUAAUAUUGCACAGAU .(((.........(((((........)))))-..(((........-..-........))).))).. ( -5.87, z-score = -2.18, R) >droYak2.chr2R 2711786 60 + 21139217 CCUGUUAACUACACUUUGAUAACAACCAAAG-AAGUACAAGACAA-CG-AAUAAUUUUUCACA--- ..((((...(((.(((((........)))))-..)))...)))).-.(-((......)))...--- ( -6.70, z-score = -1.55, R) >droEre2.scaffold_4845 9783059 58 + 22589142 CCUGUUAACUAAACUUUGACAACGACCAAAG-AAGUACAAGACAA-UG-CAUAAUUUUUCA----- ..((((.(((...(((((........)))))-.)))....)))).-..-............----- ( -4.10, z-score = 0.21, R) >dp4.chr3 16639912 64 - 19779522 CCUGUUCUAUAAACUUUGAUGGUUACCAAAGUAAAUGCAAGACAA-CA-AACAUAUUUUCAAUUAC ..((((.(((..((((((........))))))..)))...)))).-..-................. ( -5.30, z-score = 0.49, R) >droPer1.super_4 7031616 64 + 7162766 CCUGUUCUAUAAACUUUGAUGGUUACCAAAGUAAAUGCAAGACAA-CA-AACAUUUUUUCAAUUAC ..((((.(((..((((((........))))))..)))...)))).-..-................. ( -5.30, z-score = 0.61, R) >droWil1.scaffold_181009 2764202 60 + 3585778 CCUGUGACAAAUACUUUGAUCUAAAUCAAAGUAAAUACAAGACAA-UGCAGUAAUUUUUCU----- ...........(((((((((....)))))))))............-...............----- ( -8.20, z-score = -1.20, R) >droVir3.scaffold_12875 7710806 58 - 20611582 CCUGUUCGAGAUCCUUUGAUGUAUAUCAAAGAAAAUACAAGACAA---CAAUUUUACAGUU----- .((((........(((((((....)))))))(((((.........---..)))))))))..----- ( -6.30, z-score = -0.20, R) >droMoj3.scaffold_6496 7264382 66 + 26866924 CCUGUUGCAGAUCCUUUGAUGUGCACCAAAGAACACACAAGAUAAGUGCAGUUAUUUCCUUCAGUU ..((.((((.(((....))).)))).))...(((.(((.......)))..)))............. ( -9.60, z-score = 0.38, R) >droGri2.scaffold_15245 1229998 50 - 18325388 CCUGUUACAGAUCCUUUGAUGUUCAUCAAAGCAAAUACAAGACAA---CAAUC------------- ..((((.......(((((((....))))))).........)))).---.....------------- ( -6.39, z-score = -0.93, R) >consensus CCUGUUAAAUAAACUUUGAUAACAACCAAAG_AAAUACAAGACAA_CG_AAUAAUUUUUCACA___ .............(((((........)))))................................... ( -2.91 = -2.91 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:09 2011