| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,031,468 – 13,031,588 |
| Length | 120 |
| Max. P | 0.925398 |

| Location | 13,031,468 – 13,031,588 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Shannon entropy | 0.50665 |
| G+C content | 0.56617 |
| Mean single sequence MFE | -39.97 |
| Consensus MFE | -31.21 |
| Energy contribution | -31.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
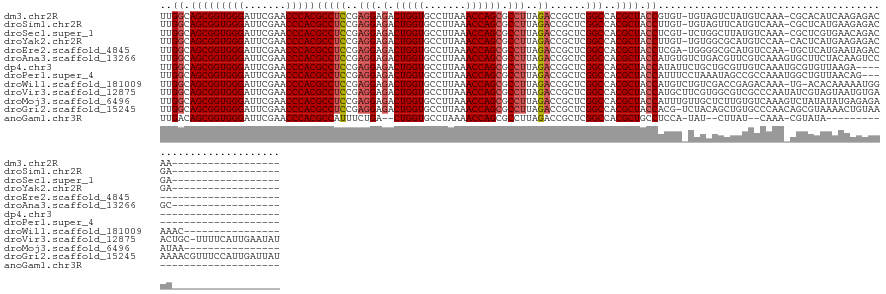
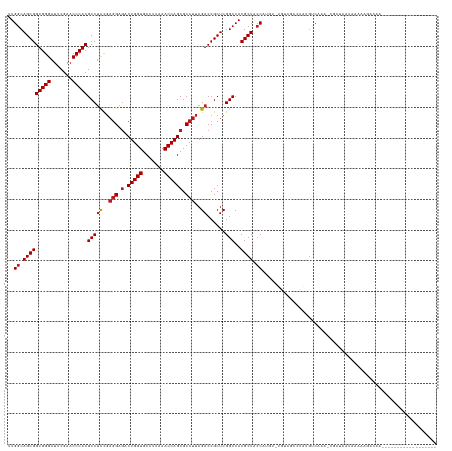
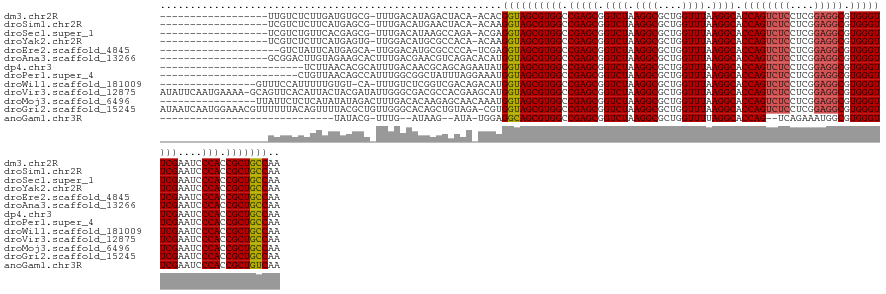
>dm3.chr2R 13031468 120 + 21146708 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCGUGU-UGUAGUCUAUGUCAAA-CGCACAUCAAGAGACAA------------------ .(((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))).(((((.....-.)))))...((((...-............)))).------------------ ( -37.36, z-score = -0.22, R) >droSim1.chr2R 11770120 120 + 19596830 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCUUGU-UGUAGUUCAUGUCAAA-CGCUCAUGAAGAGACGA------------------ .(((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))).(((((.....-.)))))..........-(((((.....))).)).------------------ ( -39.50, z-score = -0.72, R) >droSec1.super_1 10546954 120 + 14215200 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCUCGU-UCUGGCUUAUGUCAAA-CGCUCGUGAACAGACGA------------------ ..((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).))((((-.(((..(((((.....-....))))).)))))))------------------ ( -41.00, z-score = -0.65, R) >droYak2.chr2R 2705873 120 - 21139217 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCUUGU-UGUGGCGCAUGUCCAA-CACUCAUGAAGAGACGA------------------ .(((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..))))((((((.....-.))))))..(((((..-((....))..).)))).------------------ ( -41.70, z-score = -0.86, R) >droEre2.scaffold_4845 9777242 118 - 22589142 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCUCGA-UGGGGCGCAUGUCCAA-UGCUCAUGAAUAGAC-------------------- ((((((((.(((((.......)))))((((((((((.(.(((((.......))))))(((...........))).......))))).-.))))))).)).))))-...............-------------------- ( -41.00, z-score = -0.51, R) >droAna3.scaffold_13266 6314087 122 - 19884421 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAUGUGUCUGACGUUCGUCAAAGUGCUUCUACAAGUCCGC------------------ ..((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))((((......)))).((((....))))..(..(((....)))..)..------------------ ( -38.90, z-score = 0.06, R) >dp4.chr3 16625160 116 + 19779522 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAUAUUCUGCUGCGUUGUCAAAUGCGUGUUAAGA------------------------ .(((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).)))...(((((((((((....)))))).))..)))------------------------ ( -39.50, z-score = -0.77, R) >droPer1.super_4 7025894 117 - 7162766 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAUUUCCUAAAUAGCCGCCAAAUGGCUGUUAACAG----------------------- .(((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).))).......((((((((.....)))))))).....----------------------- ( -42.50, z-score = -2.36, R) >droWil1.scaffold_181009 2752782 122 - 3585778 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAUGUCUGUCGACCGAGACAAA-UG-ACACAAAAAUGGAAAC---------------- .(((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).)))((((((((......))))..-.)-))).............---------------- ( -40.80, z-score = -2.08, R) >droVir3.scaffold_12875 12905539 139 - 20611582 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAUGCUUCGUGGCGUCGCCCAAUAUCGUAGUAAUGUGAACUGC-UUUUCAUUGAAUAU ((((.(((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).)))))(((.(((((((........))))))).))))))....(((((.......)))))-.............. ( -46.00, z-score = -1.03, R) >droMoj3.scaffold_6496 7252999 124 - 26866924 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAUUUGUUGCUCUUGUGUCAAAGUCUAUAUAUGAGAGAAUAA---------------- .(((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).))).(((((.((((..(((..........)))..)))))))))---------------- ( -41.50, z-score = -1.77, R) >droGri2.scaffold_15245 1223364 139 + 18325388 UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCACG-UCUACAGCUGUGCCCAACAGCGUAAAACUGUAAAAAACGUUUCCAUUGAUUAU .(((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).)))..-..(((.(((((.....))))))))............................. ( -42.00, z-score = -0.92, R) >anoGam1.chr3R 39590462 103 - 53272125 UUGACAGCGGUGGGAUUCGAACCCACGCCAUUUCUGA--CUGGUGCCUAAAACCAGCGCCUUAGACCGCUCGGCCACGCUGCCUCCA-UAU--CUUAU--CAAA-CGUAUA----------------------------- ....((((((((((.......)))))(((...(((((--..(((((.........))))))))))......)))..)))))......-...--.....--....-......----------------------------- ( -27.80, z-score = -1.16, R) >consensus UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAUGU_UGUCGCCUAUGUCAAA_CGCUCAUAAAAAGACAA__________________ ..((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).))......................................................... (-31.21 = -31.52 + 0.31)
| Location | 13,031,468 – 13,031,588 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Shannon entropy | 0.50665 |
| G+C content | 0.56617 |
| Mean single sequence MFE | -44.17 |
| Consensus MFE | -37.90 |
| Energy contribution | -38.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
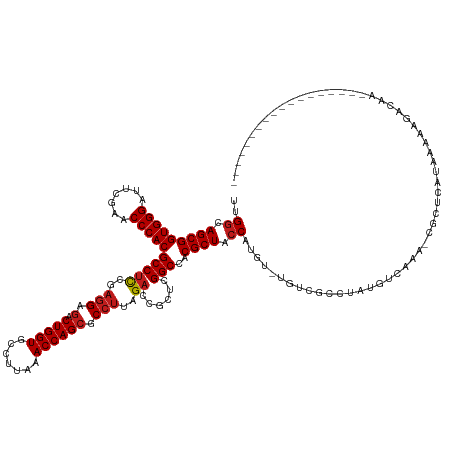
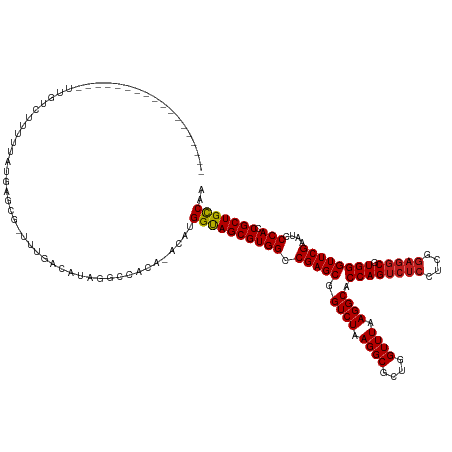
>dm3.chr2R 13031468 120 - 21146708 ------------------UUGUCUCUUGAUGUGCG-UUUGACAUAGACUACA-ACACGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ------------------........((.((((.(-((((...)))))))))-.)).((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).. ( -42.90, z-score = -0.57, R) >droSim1.chr2R 11770120 120 - 19596830 ------------------UCGUCUCUUCAUGAGCG-UUUGACAUGAACUACA-ACAAGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ------------------..((...((((((....-.....))))))..)).-....((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).. ( -42.90, z-score = -0.87, R) >droSec1.super_1 10546954 120 - 14215200 ------------------UCGUCUGUUCACGAGCG-UUUGACAUAAGCCAGA-ACGAGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ------------------(((((((.......((.-..........))))).-))))((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).. ( -43.61, z-score = -0.33, R) >droYak2.chr2R 2705873 120 + 21139217 ------------------UCGUCUCUUCAUGAGUG-UUGGACAUGCGCCACA-ACAAGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ------------------....(((.....)))((-(((...........))-))).((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).. ( -44.30, z-score = -0.48, R) >droEre2.scaffold_4845 9777242 118 + 22589142 --------------------GUCUAUUCAUGAGCA-UUGGACAUGCGCCCCA-UCGAGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA --------------------(((((..........-.)))))..........-....((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).. ( -43.00, z-score = -0.06, R) >droAna3.scaffold_13266 6314087 122 + 19884421 ------------------GCGGACUUGUAGAAGCACUUUGACGAACGUCAGACACAUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ------------------((............))..((((((....))))))....(((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))). ( -45.60, z-score = -0.62, R) >dp4.chr3 16625160 116 - 19779522 ------------------------UCUUAACACGCAUUUGACAACGCAGCAGAAUAUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ------------------------.........((..........)).........(((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))). ( -40.30, z-score = -0.50, R) >droPer1.super_4 7025894 117 + 7162766 -----------------------CUGUUAACAGCCAUUUGGCGGCUAUUUAGGAAAUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA -----------------------(((......(((....))).......)))....(((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))). ( -45.02, z-score = -0.93, R) >droWil1.scaffold_181009 2752782 122 + 3585778 ----------------GUUUCCAUUUUUGUGU-CA-UUUGUCUCGGUCGACAGACAUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ----------------............((((-(.-..((((......)))))))))((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).. ( -46.40, z-score = -1.80, R) >droVir3.scaffold_12875 12905539 139 + 20611582 AUAUUCAAUGAAAA-GCAGUUCACAUUACUACGAUAUUGGGCGACGCCACGAAGCAUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ..............-(((((.......)))......((((((...))).))).)).(((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))). ( -46.00, z-score = -0.17, R) >droMoj3.scaffold_6496 7252999 124 + 26866924 ----------------UUAUUCUCUCAUAUAUAGACUUUGACACAAGAGCAACAAAUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA ----------------.................(.((((......)))))......(((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))). ( -41.40, z-score = -1.28, R) >droGri2.scaffold_15245 1223364 139 - 18325388 AUAAUCAAUGGAAACGUUUUUUACAGUUUUACGCUGUUGGGCACAGCUGUAGA-CGUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA .(((..((((....))))..)))..((((.(((((((.....))))).)))))-).(((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))). ( -52.70, z-score = -1.63, R) >anoGam1.chr3R 39590462 103 + 53272125 -----------------------------UAUACG-UUUG--AUAAG--AUA-UGGAGGCAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUUAGGCACCAG--UCAGAAAUGGCGUGGGUUCGAAUCCCACCGCUGUCAA -----------------------------......-....--.....--...-....((((((((((.(((((.(((((((((....))))))))).((((--(((....)))).))))))))....))).))))))).. ( -40.10, z-score = -2.57, R) >consensus __________________UUGUCUUUUUAUGAGCG_UUUGACAUAGGCCACA_ACAUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA .........................................................((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).. (-37.90 = -38.07 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:07 2011