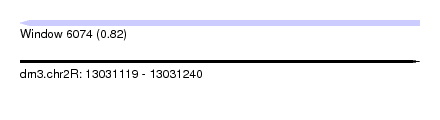
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,031,119 – 13,031,240 |
| Length | 121 |
| Max. P | 0.821776 |

| Location | 13,031,119 – 13,031,240 |
|---|---|
| Length | 121 |
| Sequences | 14 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Shannon entropy | 0.40557 |
| G+C content | 0.42904 |
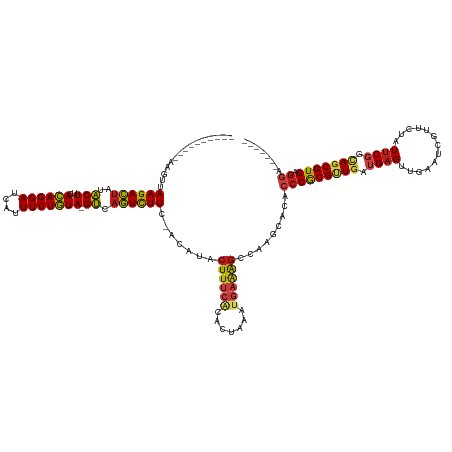
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
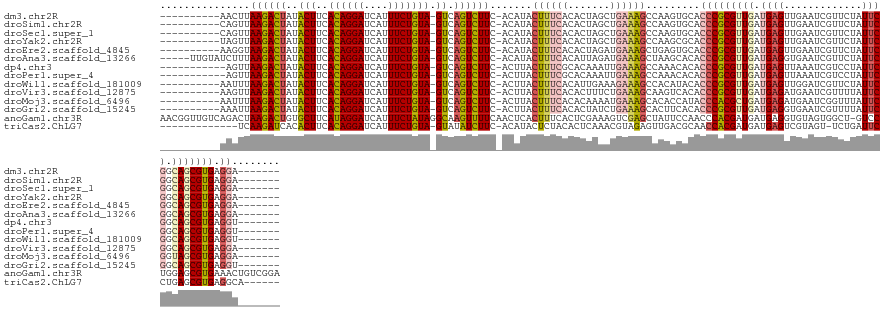
>dm3.chr2R 13031119 121 - 21146708 ----------AACUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACAUACUUUCACACUAGCUGAAAGCCAAGUGCACCCGCGUUGAUGAGUUGAAUCGUUCUAUUCGGCAGCGUGAGGA------- ----------.((((((((((..(((..((((((....)))))))-)).)))))).-.....((((((.......))))))..))))...(((((((((.(((((.((.....)).))))).))))))).)).------- ( -32.80, z-score = -1.39, R) >droSim1.chr2R 11769771 121 - 19596830 ----------CAGUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACAUACUUUCACACUAGCUGAAAGCCAAGUGCACCCGCGUUGAUGAGUUGAAUCGUUCUAUUCGGCAGCGUGAGGA------- ----------..((.((((((..(((..((((((....)))))))-)).)))))).-)).........((((.((.....))..))))..(((((((((.(((((.((.....)).))))).))))))).)).------- ( -34.10, z-score = -1.60, R) >droSec1.super_1 10546600 121 - 14215200 ----------CAGUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACAUACUUUCACACUAGCUGAAAGCCAAGUGCACCCGCGUUGAUGAGUUGAAUCGUUCUAUUCGGCAGCGUGAGGA------- ----------..((.((((((..(((..((((((....)))))))-)).)))))).-)).........((((.((.....))..))))..(((((((((.(((((.((.....)).))))).))))))).)).------- ( -34.10, z-score = -1.60, R) >droYak2.chr2R 2705498 121 + 21139217 ----------UAGUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACAUACUUUCACACUAGCUGAAAGCCAAGCGCACCCGCGUUGAUGAGUUGAAUCGUUCUAUUCGGCAGCGUGAGGA------- ----------..((.((((((..(((..((((((....)))))))-)).)))))).-))....((((((.((.((((((.....(((((....)))))(((((......)))))...)))))))).)))))).------- ( -36.10, z-score = -2.32, R) >droEre2.scaffold_4845 9776881 121 + 22589142 ----------AAGGUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACAUACUUUCACACUAGAUGAAAGCUGAGUGCACCCGCGUUGAUGAGUUGAAUCGUUCUAUUCGGCAGCGUGAGGA------- ----------.....((((((..(((..((((((....)))))))-)).))))))(-((.(((((((((....).)))))).)).)))..(((((((((.(((((.((.....)).))))).))))))).)).------- ( -33.40, z-score = -0.81, R) >droAna3.scaffold_13266 6313714 126 + 19884421 -----UUGUAUCUUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACAUACUUUCACAUUAGAUGAAAGCUAAGCACACCCGCGUUGAUGAGGUGAAUCGUUCUAUUCGGCAGCGUGAGGA------- -----.(((......((((((..(((..((((((....)))))))-)).)))))).-.....(((((((....).))))))....)))..(((((((((.((((..((.....))..)))).))))))).)).------- ( -29.80, z-score = -0.41, R) >dp4.chr3 16624784 120 - 19779522 -----------AGUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACUUACUUUCGCACAAAUUGAAAGCCAAACACACCCGCGUUGAUGAGUUAAAUCGUCCUAUUCGGCAGCGUGAGGU------- -----------(((.((((((..(((..((((((....)))))))-)).)))))).-)))..((((((.......))))))........((((((((((.(((((...........))))).))))))).)))------- ( -31.50, z-score = -2.39, R) >droPer1.super_4 7025518 120 + 7162766 -----------AGUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACUUACUUUCGCACAAAUUGAAAGCCAAACACACCCGCGUUGAUGAGUUAAAUCGUCCUAUUCGGCAGCGUGAGGU------- -----------(((.((((((..(((..((((((....)))))))-)).)))))).-)))..((((((.......))))))........((((((((((.(((((...........))))).))))))).)))------- ( -31.50, z-score = -2.39, R) >droWil1.scaffold_181009 2752413 121 + 3585778 ----------AAUUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACUUACUUUCACAUUGAAAGAAAGCCACAUACACCCGCGUUGAUGAGUUGGAUCGUUCUAUUCGGCAGCGUGAGGU------- ----------.....((((((..(((..((((((....)))))))-)).)))))).-.(((.(((((.....))))).)))........((((((((((.(((((.(((....)))))))).))))))).)))------- ( -34.50, z-score = -2.79, R) >droVir3.scaffold_12875 7704088 121 - 20611582 ----------AAGUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACUUACUUUCACACUUUCUGAAAGCAAGUCACACCCGCGUUGAUGAGAUGAAUCGUUUUAUUCGGCAGCGUGAGGA------- ----------((((.((((((..(((..((((((....)))))))-)).)))))).-)))).((((((.......)))))).........(((((((((.((((.((((....)))))))).))))))).)).------- ( -34.00, z-score = -2.28, R) >droMoj3.scaffold_6496 7252608 121 + 26866924 ----------AAUUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACUUACUUUCACACAAAAUGAAAGCACACCAUACCCACGCUGAUGAGAUGAAUCGGUUUAUUCGGUAGCGUGAGGA------- ----------.....((((((..(((..((((((....)))))))-)).)))))).-.....((((((.......)))))).........(((((((((.((((.((((....)))))))).))))))).)).------- ( -30.60, z-score = -2.29, R) >droGri2.scaffold_15245 1222974 121 - 18325388 ----------AAAUUAAGACUAUACUUCACAGGAUCAUUUCUGUA-GUCAGUCUUC-ACUUACUUUCACACUAUCUGAAAGCACUUCACACCCGCGUUGAUGAGGUGAAUCGUUUUAUUCGGCAGCGUGAGGU------- ----------.....((((((..(((..((((((....)))))))-)).)))))).-.....((((((.......))))))........((((((((((.((((.((((....)))))))).))))))).)))------- ( -30.50, z-score = -1.73, R) >anoGam1.chr3R 26805074 139 + 53272125 AACGGUUGUCAGACUAAGACUGUGCUUCAUAGGAUCAUUUCUAUAGGCAAGUUUUCAACUCACUUUCACUCGAAAGUCGAGCUAUUCCAACCCACGAUGAUGAGGUGUAGUGGCU-GUCCUGGAGCGUGAAACUGUCGGA .((((((.(((....((((((.(((((.((((((....)))))))))))))))))...(((((((((....))))))..(((((((...(((((......)).)))..)))))))-......)))..))))))))).... ( -35.90, z-score = 0.60, R) >triCas2.ChLG7 9949540 118 - 17478683 -------------UCAAGAUCACACUUCACAGGAUCAUUUCUGUA-GUAUAUCUUC-ACAUACUCUACACUCAAACGUAGAGUUGACGCAACCACGAUGAUGAGUCGUAGU-UCUGAUUCCUGAGCGUGAGGCA------ -------------..(((((...(((..((((((....)))))))-))..))))).-.((.(((((((........)))))))))..((...((((.....((((((....-..)))))).....))))..)).------ ( -29.50, z-score = -0.59, R) >consensus __________AAGUUAAGACUAUACUUCACAGGAUCAUUUCUGUA_GUCAGUCUUC_ACAUACUUUCACACUAAAUGAAAGCCAAGCACACCCGCGUUGAUGAGUUGAAUCGUUCUAUUCGGCAGCGUGAGGA_______ ...............((((((..((...((((((....))))))..)).)))))).......((((((.......)))))).........(((((((((.((((.............)))).))))))).))........ (-21.27 = -21.74 + 0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:05 2011