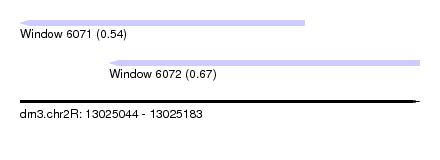
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,025,044 – 13,025,183 |
| Length | 139 |
| Max. P | 0.672171 |

| Location | 13,025,044 – 13,025,143 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 69.92 |
| Shannon entropy | 0.52242 |
| G+C content | 0.52456 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -16.66 |
| Energy contribution | -18.38 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13025044 99 - 21146708 AUACGGUUGACGACCUCCUGAUGGUUGAUUAUCUGGAUAAUACGAAGGAGGCGGUGGAAAACGACCGAAGACAAAGGGAGGUGGUCCAAUGGAUCCUGG------ ...(.((((((.(((((((..((.((((((((....))))).)))......((((.(....).))))....))..))))))).)).)))).).......------ ( -29.30, z-score = -1.16, R) >droSim1.chr2R 11763460 99 - 19596830 AAACGGUUGACGACCUCCAGAUGGUUGGUUAGCUGGAUAAUACGAAGGAGACGGUGGAAAACGACCGAAGGCAAAGGGGGGUGGUCCAUUGGAUCCUGG------ ...((((((((((((.......)))).))))))))..........((((..(((((((..((..((..........))..))..)))))))..))))..------ ( -34.90, z-score = -2.45, R) >droSec1.super_1 10540391 99 - 14215200 AAACGGUUGACGACCUCCAGAUGGUUGGUUAUCUGGAUAAUACGAAGGAGGCGGUGGAAAACGACCAAAGGCAAAGGGGGGUGGUCCAUUGGAUCCUGG------ ....((((...))))(((((((((....)))))))))........((((..(((((((..((..((..........))..))..)))))))..))))..------ ( -31.70, z-score = -1.27, R) >droEre2.scaffold_4845 9770530 105 + 22589142 AUACGGUUGACGACCUCCAGAUGGUUGAUUUUCUGGAUAAUACGGAGGGGGCGGUGGGAAGCGACCGAAGGCAAAGGGAGGUGGUCCAGCGGAUCCUGGGCCAAG ...((.....))((((((.....(((..(((((((.......)))))))..((((.(....).))))..)))....))))))(((((((......)))))))... ( -36.80, z-score = -1.14, R) >droAna3.scaffold_13266 6306885 84 + 19884421 AUCUGGGCGAUAUCCUCCUGGAACUUGAUUAUAGGGAUUUCUCGGAAAAUAUCCUG----UUAGCUGAGGCU-----GUGGCACUCCAACUGG------------ ...(((((.(((.((((....(((.........(((....)))(((.....))).)----))....)))).)-----)).))...))).....------------ ( -17.80, z-score = 1.10, R) >consensus AUACGGUUGACGACCUCCAGAUGGUUGAUUAUCUGGAUAAUACGAAGGAGGCGGUGGAAAACGACCGAAGGCAAAGGGAGGUGGUCCAAUGGAUCCUGG______ ....(((.....)))(((((((((....)))))))))........((((..((.((((..((..((..........))..))..)))).))..))))........ (-16.66 = -18.38 + 1.72)
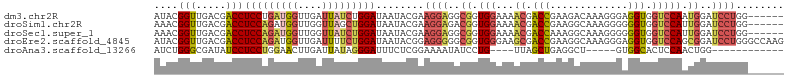
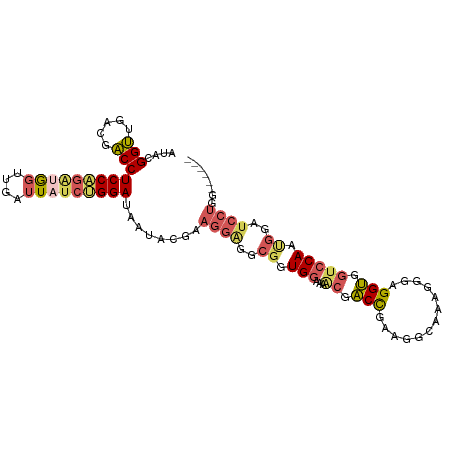
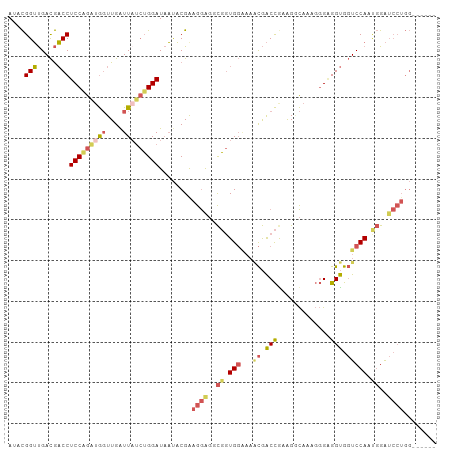
| Location | 13,025,075 – 13,025,183 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.50652 |
| G+C content | 0.51098 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -19.74 |
| Energy contribution | -21.97 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13025075 108 - 21146708 GACUGCCAGGUUGUCCUUGAACUGCACCGACGUUCGGAUAAUACGGUUGACGACCUCCUGAUGGUUGAUUAUCUGGAUAAUACGAAGGAGGCGGUGGAAAACGACCGA ........((((((..((..(((((.((..((((((((((((........(((((.......)))))))))))))).....)))..))..))))).))..)))))).. ( -31.10, z-score = -0.85, R) >droSim1.chr2R 11763491 108 - 19596830 GACUGCCAGGUUGUCCUUGAACUGCACCACCAUUCGGAUAAAACGGUUGACGACCUCCAGAUGGUUGGUUAGCUGGAUAAUACGAAGGAGACGGUGGAAAACGACCGA ........((((((...((.....))(((((.((((.......((((((((((((.......)))).)))))))).......))))(....))))))...)))))).. ( -34.44, z-score = -1.92, R) >droSec1.super_1 10540422 108 - 14215200 GACUGCCAGGUUGUCCAUGAACUGCACCACCGUUUGGAUAAAACGGUUGACGACCUCCAGAUGGUUGGUUAUCUGGAUAAUACGAAGGAGGCGGUGGAAAACGACCAA .((((((..((((((((.((.....(((((((((((((......((((...))))))))))))).))))..)))))))))).(....).))))))((.......)).. ( -36.80, z-score = -2.28, R) >droYak2.chr2R 2698551 108 + 21139217 GACCGCCAGGUUGUCCGUGAACUGCACCACCGUUCGGAUAAUACGGUUGACGACCUCCAGAUGAUUGAUUAUCUGGGUAAGAGGGAGGAGGCGGUGGGAAACGACCGA ..((.((((((((((.(((.....))).(((((.........))))).))))))))((((((((....)))))))).......)).))...((((.(....).)))). ( -41.30, z-score = -2.20, R) >droEre2.scaffold_4845 9770567 108 + 22589142 GACCGCCAGCUUGUCCGUGAACUGCACCACCGUUCGGAUAAUACGGUUGACGACCUCCAGAUGGUUGAUUUUCUGGAUAAUACGGAGGGGGCGGUGGGAAGCGACCGA .((((((..((..((((((.......(.(((((.........))))).)......((((((..........))))))...)))))))).))))))((.......)).. ( -33.60, z-score = 0.32, R) >droAna3.scaffold_13266 6306905 89 + 19884421 ---------GAUUUCCA-AAGUUUCCUGGCUGUU-----GAUCUGGGCGAUAUCCUCCUGGAACUUGAUUAUAGGGAUUUCUCGGAAAAUAUCCUGUUAGCUGA---- ---------..(((((.-.((.((((((...((.-----.(((..((.(.....).))..))..)..))..))))))...)).)))))................---- ( -18.70, z-score = 0.80, R) >consensus GACUGCCAGGUUGUCCAUGAACUGCACCACCGUUCGGAUAAUACGGUUGACGACCUCCAGAUGGUUGAUUAUCUGGAUAAUACGAAGGAGGCGGUGGAAAACGACCGA .((((((.(((((((....(((((.....((....))......))))))))))))(((((((((....)))))))))............))))))............. (-19.74 = -21.97 + 2.23)

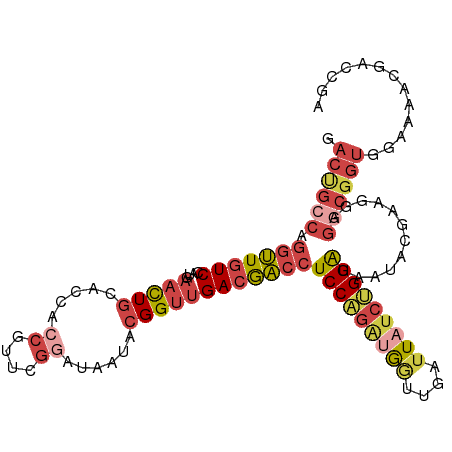

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:03 2011