| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,022,715 – 13,022,806 |
| Length | 91 |
| Max. P | 0.900444 |

| Location | 13,022,715 – 13,022,806 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 62.27 |
| Shannon entropy | 0.52860 |
| G+C content | 0.33260 |
| Mean single sequence MFE | -17.27 |
| Consensus MFE | -9.73 |
| Energy contribution | -8.73 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13022715 91 + 21146708 UGAUCCAGCAUUUGCUGCAAAGGAUUAGGGAAACCCAG-UAAAAUUUAUCAGAUCUAUAAAUA-UUAGUAAUAGAUUUACGAAAUUAAUGAUA ((((((.(((.....)))...))))))((....))...-...(((((...((((((((.....-......))))))))...)))))....... ( -16.80, z-score = -1.71, R) >droMoj3.scaffold_6442 49846 82 + 113228 UGGUCCAACAUUCGUUGCGACGGAUUGGGGAAACCCAGACAAAACCCAACCAGACCAUUCUUAAUAAUACUAAUAUU------AUUUA----- (((((.....(((((....)))))(((((....)))))..............))))).....(((((((....))))------)))..----- ( -19.80, z-score = -2.48, R) >droGri2.scaffold_14906 42973 87 - 14172833 UGGUCCAGCACUUGCUGCUAGGGAUUAGGAAAACCUAG-UGAAAUCUAAUUGAACAAUUUAUAGUUAGAGAUAUGUUUAC---AUUAAAUA-- ..((((((((.....))))..))))((((....))))(-((((.((((((((.........))))))))......)))))---........-- ( -15.20, z-score = -0.14, R) >consensus UGGUCCAGCAUUUGCUGCAAAGGAUUAGGGAAACCCAG_UAAAAUCUAACAGAACAAUUAAUA_UUAGAAAUAUAUUUAC___AUUAA__A__ (((..((((....)))).......(((((....))))).......)))............................................. ( -9.73 = -8.73 + -0.99)

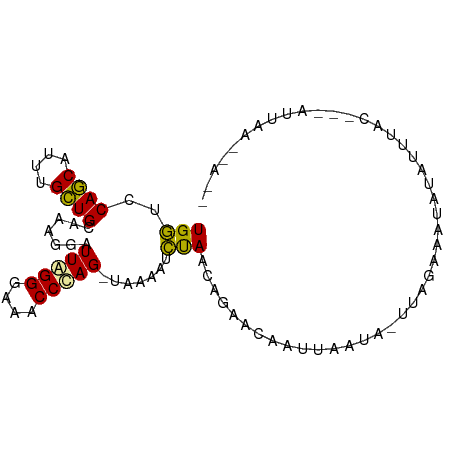
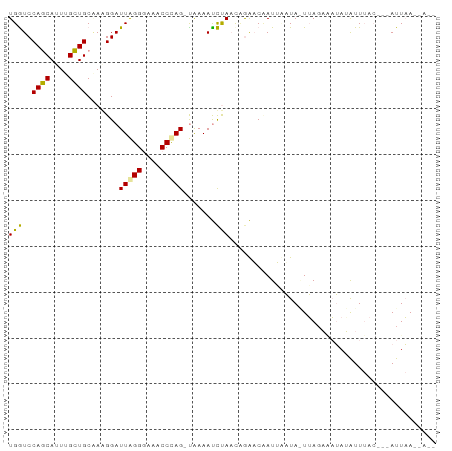
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:01 2011