| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,021,455 – 13,021,589 |
| Length | 134 |
| Max. P | 0.688990 |

| Location | 13,021,455 – 13,021,561 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Shannon entropy | 0.37789 |
| G+C content | 0.47312 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -22.33 |
| Energy contribution | -23.55 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
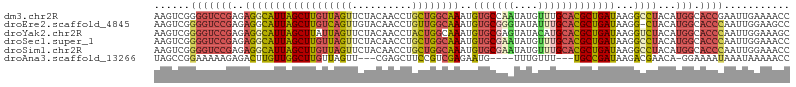
>dm3.chr2R 13021455 106 - 21146708 AAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCCAAUAUGUUUGCACGCUGAUAAGGCCUACAUGGCACCGAAUUGAAAACC ...((((.(((....((((((((((((((((((..........))))))))..((((.((....)).))))))))))...))))....))).)))).......... ( -31.70, z-score = -1.01, R) >droEre2.scaffold_4845 9768116 105 + 22589142 AAGUCGGGGUCCGAGAGGCAUUAGCUUGUCAGUUGUACAACCUGUUGGCAAAUGUGCGGGUAUAUUUGCACGCUGAUAAGG-CUACAUGGCACCCAAUUGGAAGCC .....((..(((((..(((((((((((((((((.((.(((....)))(((((((((....))))))))))))))))))).)-))).)))....))..)))))..)) ( -34.00, z-score = -0.80, R) >droYak2.chr2R 2696385 106 + 21139217 AAGUCGGGGUCCGAGAGGCAUUAGCUUAUUAGUUCUACAACCUACUGGCAAAUGUGCGAGUAUACAUGCACGCUGAUAAGGUCUACAUGGCACCCAAUUGGAAAGC ......(((((((.(((((.....(((((((((.........((((.(((....))).)))).........))))))))))))).).))).))))........... ( -27.97, z-score = -0.33, R) >droSec1.super_1 10537093 106 - 14215200 AAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCGAAUAUGUUUGCACGCUGAUAAGGCCUACAUGGCACCCAAUUGGAAACC ......(((((((.(((((((((((((((((((..........))))))))..(((((((....)))))))))))))...)))).).))).))))....(....). ( -37.10, z-score = -2.31, R) >droSim1.chr2R 11760203 106 - 19596830 AAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCGAAUAUGUUUGCACGCUGAUAAGGCCUACAUGGCACCCAAUUGGAAACC ......(((((((.(((((((((((((((((((..........))))))))..(((((((....)))))))))))))...)))).).))).))))....(....). ( -37.10, z-score = -2.31, R) >droAna3.scaffold_13266 6304362 95 + 19884421 UAGCCGGAAAAAGAGACUUGUUGGCUUGUUAGUU---CGAGCUUCCGUCGAGAAUG----UUUGUUU---UGCCGAUAAGACGAACA-GGAAAAUAAAUAAAAACC ...((.........(((..(..((((((......---))))))..)))).....((----(((((((---((....)))))))))))-))................ ( -21.00, z-score = -1.08, R) >consensus AAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCGAAUAUGUUUGCACGCUGAUAAGGCCUACAUGGCACCCAAUUGGAAACC ......(((((((..((((((((((((((((((..........))))))))..(((((((....)))))))))))))...))))...))).))))........... (-22.33 = -23.55 + 1.22)
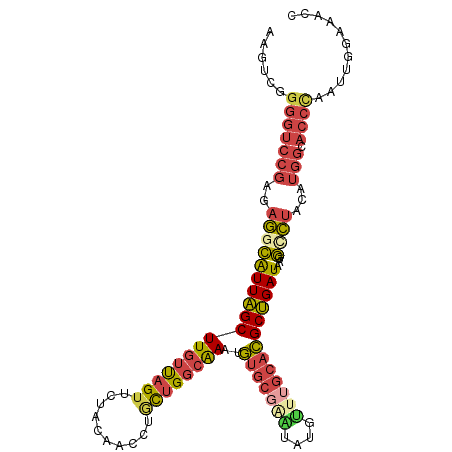
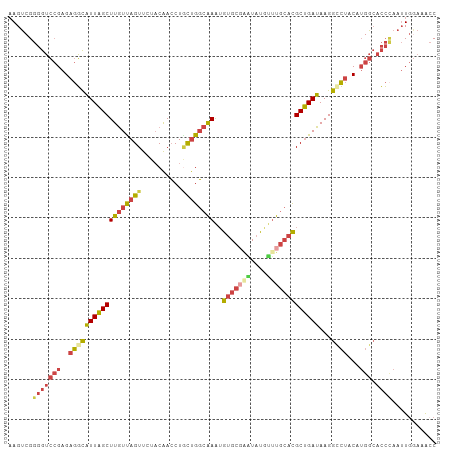
| Location | 13,021,493 – 13,021,589 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Shannon entropy | 0.40803 |
| G+C content | 0.47068 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13021493 96 - 21146708 UCAGUUUCGGAUCCACUUCG------------AGAGGCAGAAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCCAAUAUGUUUG ....(((((((((((((((.------------.......)))))..))))))))))(((((....((((((((..........))))))))..))))).......... ( -34.30, z-score = -2.79, R) >droEre2.scaffold_4845 9768153 96 + 22589142 UCAGUUUGGGAUCCACUUCG------------AGAGGCAGAAGUCGGGGUCCGAGAGGCAUUAGCUUGUCAGUUGUACAACCUGUUGGCAAAUGUGCGGGUAUAUUUG .(((.((((((((((((((.------------.......)))))..))))))((.((((....)))).)).......))).)))....((((((((....)))))))) ( -29.80, z-score = -1.36, R) >droYak2.chr2R 2696423 101 + 21139217 UCAGUUUCGGAUCCACUUCGUGA-------GAAGAAGCAGAAGUCGGGGUCCGAGAGGCAUUAGCUUAUUAGUUCUACAACCUACUGGCAAAUGUGCGAGUAUACAUG ....((((((((((.((((....-------)))).....(....).))))))))))(((....)))................((((.(((....))).))))...... ( -29.50, z-score = -1.68, R) >droSec1.super_1 10537131 96 - 14215200 UCAGUUUCGGAUCCACUUCG------------AGAGGCAGAAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCGAAUAUGUUUG ((..(((((((((((((((.------------.......)))))..)))))))))).((((....((((((((..........))))))))..))))))......... ( -30.90, z-score = -1.66, R) >droSim1.chr2R 11760241 96 - 19596830 UCAGUUUCGGAUCCACUUCG------------AGAGGCAGAAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCGAAUAUGUUUG ((..(((((((((((((((.------------.......)))))..)))))))))).((((....((((((((..........))))))))..))))))......... ( -30.90, z-score = -1.66, R) >droAna3.scaffold_13266 6304397 100 + 19884421 UCAGUUUCAGAUUCAUUUAGAAAAUCAUUCAGAGAGACAGUAGCCGGAAAAAGAGACUUGUUGGCUUGUUAGUUCGAGCUUCCGUCGA-GAAUGUUUGUUU------- ....((((.((.....)).))))..(((((.....(((.(.(((((((....(((.((....))))).....)))).))).).)))..-))))).......------- ( -16.50, z-score = 1.59, R) >consensus UCAGUUUCGGAUCCACUUCG____________AGAGGCAGAAGUCGGGGUCCGAGAGGCAUUAGCUUGUUAGUUCUACAACCUGCUGGCAAAUGUGCGAAUAUGUUUG ....(((((((((((((((....................)))))..)))))))))).((((....((((((((..........))))))))..))))........... (-19.33 = -19.87 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:29:00 2011