
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,983,611 – 12,983,706 |
| Length | 95 |
| Max. P | 0.620512 |

| Location | 12,983,611 – 12,983,706 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.56140 |
| G+C content | 0.50629 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -12.35 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620512 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
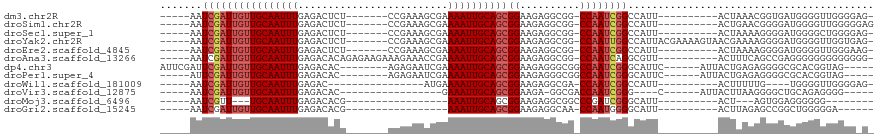
>dm3.chr2R 12983611 95 - 21146708 -----AAUCGAUUGUUGCAAUUUGAGACUCU-------CCGAAAGCGAAAAUUGCAGCGGAAGAGGCGG-CCAAUCGGCCAUU----------ACUAAACGGUGAUGGGGUUGGGGAG- -----..((..((((((((((((..(.((..-------.....)))..))))))))))))..)).....-((((((..(((((----------(((....))))))))))))))....- ( -30.10, z-score = -1.65, R) >droSim1.chr2R 11721202 96 - 19596830 -----AAUCGAUUGUUGCAAUUUGAGACUCU-------CCGAAAGCGAAAAUUGCAGCGGAAGAGGCGG-CCAAUCGGCCAUU----------ACUGAACGGGGAUGGGGUUGGGGGAG -----..((..((((((((((((..(.((..-------.....)))..))))))))))))..)).....-((((((..((((.----------.((....))..))))))))))..... ( -27.40, z-score = -0.36, R) >droSec1.super_1 10494230 95 - 14215200 -----AAUCGAUUGUUGCAAUUUGAGACUCU-------CCGAAAGCGAAAAUUGCAGCGGAAGAGGCGG-CCAAUCGGCCAUU----------ACUAAAAGGGGAUGGGGCUGGGGAG- -----..((..((((((((((((..(.((..-------.....)))..))))))))))))..))..(((-((......((((.----------.((....))..))))))))).....- ( -26.10, z-score = -0.18, R) >droYak2.chr2R 2656172 105 + 21139217 -----AAUCGAUUGUUGCAAUUUGAGACUCU-------CCGAAAGCGAAAAUUGCAGCGGAAGAGGCGG-CCAAUUGGCCAUUACGAAAAGUAACGAAAAGGGGAUGGGGUUGGUGAG- -----......(((((((..((((...((((-------(((...((((...))))..))).))))..((-((....))))....))))..))))))).....................- ( -28.10, z-score = -1.11, R) >droEre2.scaffold_4845 9730142 95 + 22589142 -----AAUCGAUUGUUGCAAUUUGAGACUCU-------CCGAAAGCGAAAAUUGCAGCGGAAGAGGCGG-CCAAUCGGCCAUU----------ACUAAAAGGGGAUGGGGUUGGGAAG- -----..((..((((((((((((..(.((..-------.....)))..))))))))))))..)).....-((((((..((((.----------.((....))..))))))))))....- ( -26.70, z-score = -0.81, R) >droAna3.scaffold_13266 6265631 102 + 19884421 -----AAUCGAUUGUUGCAAUUUGAGACACAGAGAAGAAAGAAACCGAAAAUUGCAGCGGAAGAGGCGG-CCAAUCAGGCGUU----------ACUUUCAGCCGAGGGGGGGGGGGGG- -----..((..((((((((((((..(...................)..))))))))))))..))((((.-((.....))))))----------.((..(..((....))..)..))..- ( -24.11, z-score = -0.12, R) >dp4.chr3 16579571 100 - 19779522 AUUCGAUUCGAUUGUUGCAAUUUGAGACAC--------AGAGAAUCGAAAAUUGCAGCGGAAGAGGGCGGCCAAUCGGGCAUUC------AUUACUGAGAGGGGCGCACGGUAG----- ..(((.(((..((((((((((((..((...--------......))..))))))))))))..))).(((.((..((((......------....))))...)).))).)))...----- ( -30.20, z-score = -2.28, R) >droPer1.super_4 6980635 95 + 7162766 -----AUUCGAUUGUUGCAAUUUGAGACAC--------AGAGAAUCGAAAAUUGCAGCGGAAGAGGGCGGCCAAUCGGGCAUUC------AUUACUGAGAGGGGCGCACGGUAG----- -----.(((..((((((((((((..((...--------......))..))))))))))))..))).(((.((..((((......------....))))...)).))).......----- ( -29.00, z-score = -2.55, R) >droWil1.scaffold_181009 2685617 82 + 3585778 -----AAUCGAUUGUUGCAAUUUGAGAC----------------AUGAAAAUUGCAGCGGAAGAGGCGA-CCAAUCGGCCAUU----------ACUUUUUG----UGGGGUUGGGGAG- -----..((..((((((((((((.....----------------....))))))))))))..)).....-((((((..((((.----------.......)----)))))))))....- ( -23.60, z-score = -1.84, R) >droVir3.scaffold_12875 7645735 81 - 20611582 -----AAUCGAUUGUUGCAAUUUGAGACAC-----------------GAAAUUGCAGCGGAAGA-GGCGACCAAUCGGG----C------AUUACUUAAGGGGCUGCAGAGGGG----- -----..((..((((((((((((.......-----------------.))))))))))))..))-.(((.((..(..((----.------....))..)..)).))).......----- ( -20.50, z-score = -1.18, R) >droMoj3.scaffold_6496 7166369 73 + 26866924 -----AAUCGUU---UGCAAUUUGAGACACG-----------------AAAUUGCAGCGGAAGAGGCGGCCCGAUCGGGCAUU----------ACU---AGUGGAGGGGGG-------- -----..((.((---..(.............-----------------....(((..(....)..)))((((....))))...----------...---.)..)).))...-------- ( -19.30, z-score = -0.63, R) >droGri2.scaffold_15245 1171346 80 - 18325388 -----AAUCGAUUGUUGCAAUUUGAGACACG-----------------AAAUUGCAGCGGAAGAGGCAA-CCAAUGGGGCAUU----------ACUUAGAGCCGGCUGGGGGA------ -----..((..((((((((((((........-----------------))))))))))))..))..(..-(((.(((..(...----------.....)..)))..)))..).------ ( -20.50, z-score = -0.89, R) >consensus _____AAUCGAUUGUUGCAAUUUGAGACACU_______CCGAAAGCGAAAAUUGCAGCGGAAGAGGCGG_CCAAUCGGCCAUU__________ACUAAAAGGGGAUGGGGGUGGGG_G_ .......((((((((((((((((.........................))))))))))......((....))))))))......................................... (-12.35 = -12.87 + 0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:56 2011