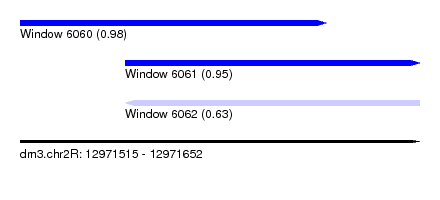
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,971,515 – 12,971,652 |
| Length | 137 |
| Max. P | 0.982667 |

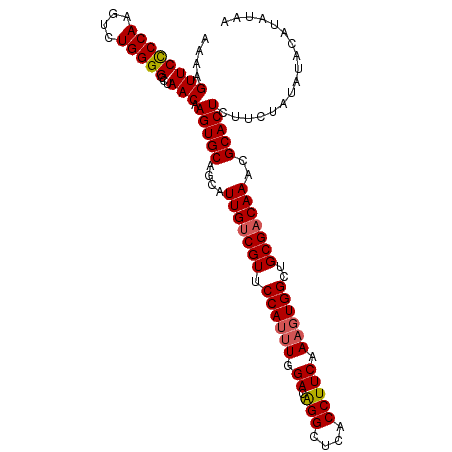
| Location | 12,971,515 – 12,971,620 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Shannon entropy | 0.10130 |
| G+C content | 0.46720 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -29.78 |
| Energy contribution | -30.06 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12971515 105 + 21146708 AAAAGUUCCCCAAGUCUGGGGCCUAACAAGUGCAGCAUUGUCGUUCCAAUUGGACGGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUAU-- ....((((((((....)))))...))).(((((....(((((((.(((.(((((.((.....)))))))..)))..)))))))..)))))...............-- ( -31.10, z-score = -2.30, R) >droEre2.scaffold_4845 9718007 107 - 22589142 AAAAGUUCUCCAAGUCUGGGGCCUAACAAGUGCAGCAUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUAUACAUAC ....((((((((....)))))...))).(((((....(((((((.((((((.((.(((....))))).))))))..)))))))..)))))................. ( -29.20, z-score = -2.13, R) >droYak2.chr2R 2644090 107 - 21139217 AAAAGUUCCCCAAGUCUGGGGCCUAACAAGUGCAGCAUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAUGUGGCUGCGCCAAACGCACUCUUCUAUAUAAAUAUAU ....((((((((....)))))...)))..(((((((...(((..(((....)))..))).(((.......))))))))))........................... ( -27.20, z-score = -1.10, R) >droSec1.super_1 10482329 107 + 14215200 AAAAGUUCCCCAAGUCUGGGGCCUAACAAGUGCAGCAUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUACUA ....((((((((....)))))...))).(((((....(((((((.((((((.((.(((....))))).))))))..)))))))..)))))................. ( -32.20, z-score = -2.67, R) >droSim1.chr2R 11709231 107 + 19596830 AAAAGUUCCCCAGGUCUGGGGCCUAACAAGUGCAGCAUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUACUA ....((((((((....)))))...))).(((((....(((((((.((((((.((.(((....))))).))))))..)))))))..)))))................. ( -32.90, z-score = -2.47, R) >consensus AAAAGUUCCCCAAGUCUGGGGCCUAACAAGUGCAGCAUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUAUAA ....((((((((....)))))...))).(((((....(((((((.((((((.((.(((....))))).))))))..)))))))..)))))................. (-29.78 = -30.06 + 0.28)

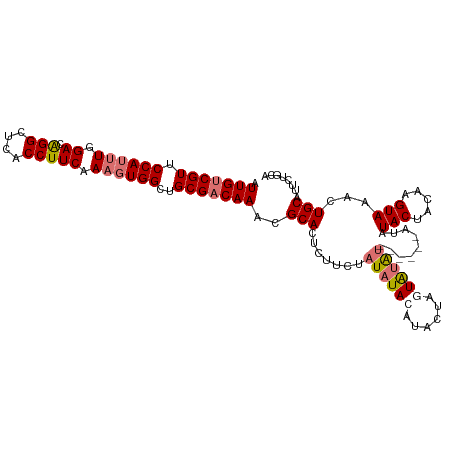
| Location | 12,971,551 – 12,971,652 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 86.52 |
| Shannon entropy | 0.22394 |
| G+C content | 0.39302 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -18.80 |
| Energy contribution | -20.93 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12971551 101 + 21146708 AUUGUCGUUCCAAUUGGACGGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUA----UGUAU------GUAUACUACAAGUAAACUGCAUUCUGCA .(((((((.(((.(((((.((.....)))))))..)))..)))))))..(((......(((((((...----.))))------)))(((.....)))...)))........ ( -25.40, z-score = -2.20, R) >droYak2.chr2R 2644126 111 - 21139217 AUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAUGUGGCUGCGCCAAACGCACUCUUCUAUAUAAAUAUAUAUACGAGUAUACACAUACUACUUGUAUACUGCAUUCUGCA .(((.(((.((((...((.(((....)))))...))))..))).)))..(((......((((((....))))))..((((((((.........))))))))......))). ( -22.90, z-score = -1.29, R) >droSec1.super_1 10482365 105 + 14215200 AUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUACUAGUAUAU------AUAUACUACAAGUAAACUGCAUUCUGCA .(((((((.((((((.((.(((....))))).))))))..)))))))..(((......(((((((......))))))------)..(((.....)))...)))........ ( -26.40, z-score = -3.15, R) >droSim1.chr2R 11709267 111 + 19596830 AUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUACUAGUAUAUAAAUAUAUAUACUACAAGUAAACUGCAUUCUGCA .(((((((.((((((.((.(((....))))).))))))..)))))))..(((..........(((....(((((((((....)))))))))...)))..........))). ( -27.95, z-score = -3.70, R) >consensus AUUGUCGUUCCAUUUGGACAGGCUCACCUUCAAAGUGGCUGCGACAAACGCACUCUUCUAUAUACAUACUAGUAUAU______AUAUACUACAAGUAAACUGCAUUCUGCA .(((((((.((((((.((.(((....))))).))))))..)))))))..(((..........(((....(((((((((....)))))))))...)))..........))). (-18.80 = -20.93 + 2.13)

| Location | 12,971,551 – 12,971,652 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.52 |
| Shannon entropy | 0.22394 |
| G+C content | 0.39302 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -20.29 |
| Energy contribution | -23.10 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12971551 101 - 21146708 UGCAGAAUGCAGUUUACUUGUAGUAUAC------AUACA----UAUGUAUAUAGAAGAGUGCGUUUGUCGCAGCCACUUUGAAGGUGAGCCCGUCCAAUUGGAACGACAAU ....(((((((.(((.((....((((((------((...----.)))))))))).))).)))))))((((..((((((.....)))).))...(((....))).))))... ( -27.80, z-score = -2.13, R) >droYak2.chr2R 2644126 111 + 21139217 UGCAGAAUGCAGUAUACAAGUAGUAUGUGUAUACUCGUAUAUAUAUUUAUAUAGAAGAGUGCGUUUGGCGCAGCCACAUUGAAGGUGAGCCUGUCCAAAUGGAACGACAAU ....((((((.(((((((.......)))))))((((.((((((....))))))...))))))))))(((...(((........)))..)))..(((....)))........ ( -27.90, z-score = -1.10, R) >droSec1.super_1 10482365 105 - 14215200 UGCAGAAUGCAGUUUACUUGUAGUAUAU------AUAUACUAGUAUGUAUAUAGAAGAGUGCGUUUGUCGCAGCCACUUUGAAGGUGAGCCUGUCCAAAUGGAACGACAAU ....(((((((.(((.((....((((((------((((....)))))))))))).))).)))))))((((...(((.((((.(((....)))...)))))))..))))... ( -29.70, z-score = -2.32, R) >droSim1.chr2R 11709267 111 - 19596830 UGCAGAAUGCAGUUUACUUGUAGUAUAUAUAUUUAUAUACUAGUAUGUAUAUAGAAGAGUGCGUUUGUCGCAGCCACUUUGAAGGUGAGCCUGUCCAAAUGGAACGACAAU ....(((((((.(((.((....(((((((((((........))))))))))))).))).)))))))((((...(((.((((.(((....)))...)))))))..))))... ( -30.60, z-score = -2.37, R) >consensus UGCAGAAUGCAGUUUACUUGUAGUAUAU______AUAUACUAGUAUGUAUAUAGAAGAGUGCGUUUGUCGCAGCCACUUUGAAGGUGAGCCUGUCCAAAUGGAACGACAAU ....(((((((.(((.((....(((((((((((........))))))))))))).))).)))))))((((..((((((.....)))).))...(((....))).))))... (-20.29 = -23.10 + 2.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:55 2011