
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,955,862 – 12,955,973 |
| Length | 111 |
| Max. P | 0.519541 |

| Location | 12,955,862 – 12,955,973 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.10 |
| Shannon entropy | 0.38257 |
| G+C content | 0.48925 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -18.15 |
| Energy contribution | -17.50 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12955862 111 - 21146708 UGGUCAUGCCCAUGGUG---------CACAACGCCAUGCCCACCUUCGAUGAUUUUGACUUCGAGGAGUUCCUCUCCUCAUUCGAGAACGAGAAUGACGACGAGCAAUUCCCGCUGUUCA ..(((((...(((((((---------.....)))))))......(((((.((........))((((((.....))))))..))))).......)))))...(((((........))))). ( -32.30, z-score = -1.39, R) >droGri2.scaffold_15245 1138313 108 - 18325388 UGGGCAUGACCAAGAUG------------CCCGUGUUGCCCACAUUUGAUGAUUUUGAUUUUGAGGAAUUUCUCUCAUCAUUCGAGAAUGAGAAUGAUGAGGAGCAAUUUCCGCUGUUUC .((((((.......)))------------)))(((.....))).....................((((.((((((((((((((........))))))))).))).)).))))........ ( -32.10, z-score = -1.77, R) >droMoj3.scaffold_6496 14370850 108 - 26866924 UGACGCUGCCCAAUAUG------------CCUGUGCUGCCCACGUUUGAUGACUUCGACUUUGAGGAAUUUCUGUCAUCAUUCGAGAACGAGAAUGAUGACGAGCAAUUUCCGCUGUUCC ....((.((.((.....------------..)).)).))....(((....)))...(((..((.((((((.((((((((((((........)))))))))).)).))))))))..))).. ( -25.60, z-score = -0.80, R) >droVir3.scaffold_12875 7610706 108 - 20611582 UGAAUUUACCCAAUAUG------------CCAGUGCUGCCCACGUUUGAUGACUUUGAUUUCGAGGAAUUUCUCUCAUCAUUCGAGAACGAUAAUGAUGAGGAGCAAUUUCCGCUGUUUC .................------------.(((((.(((.............(((((....)))))....((.(((((((((((....)))..))))))))))))).....))))).... ( -21.90, z-score = 0.15, R) >droWil1.scaffold_181009 2639865 111 + 3585778 UGGUCAUGCCCAGUC---------ACAAUAAUCUGAUGCCGAGCUUCGAUAAUUUCGAUUUUGAGGAAUUUCUCUCGUCAUUCGAGAAUGAGAAUGAUGAGGAGCAAUUUCCCUUGUUUC .((.(((...(((..---------........))))))))((((.((((.....))))......((((.((((((((((((((........))))))))).))).)).))))...)))). ( -26.60, z-score = 0.11, R) >droPer1.super_2 6410699 105 - 9036312 UGGUCAUGCCGAUG---------------GGCAACAUGCCCACCUUCGACGACUUUGACUUUGAGGAGUUUCUCUCCUCAUUCGAGAACGAGAAUGACGACGAGCAGUUCCCGCUGUUCA ..(((((..(((((---------------(((.....)))))...))).((..(((((...(((((((.....))))))).)))))..))...)))))...(((((((....))))))). ( -39.00, z-score = -3.18, R) >dp4.chr3 6195854 105 - 19779522 UGGUCAUGCCGAUG---------------GGCAACAUGCCCACCUUCGACGACUUUGACUUUGAGGAGUUUCUCUCCUCAUUCGAGAACGAGAAUGACGACGAGCAGUUCCCGCUGUUCA ..(((((..(((((---------------(((.....)))))...))).((..(((((...(((((((.....))))))).)))))..))...)))))...(((((((....))))))). ( -39.00, z-score = -3.18, R) >droAna3.scaffold_13266 6239912 120 + 19884421 UGGUUAUGCCCAUGGUGCAACAUGCCCAUAGCAACAUGCCCACCUUCGAUGACUUUGACUUCGAGGAGUUCCUCUCCUCAUUCGAAAACGAGAAUGACGACGAGCAGUUCCCCCUGUUCA .(((..(((..((((.((.....)))))).)))....))).....(((..............((((((.....)))))).((((....)))).....))).((((((......)))))). ( -32.50, z-score = -1.76, R) >droEre2.scaffold_4845 9702645 111 + 22589142 UGGUCAUGCCCAUGGUG---------CACAACGCCAUGCCCACCUUCGAUGAUUUUGACUUUGAGGAGUUCCUCUCCUCAUUCGAGAACGAGAAUGAUGACGAGCAAUUCCCGCUGUUCA ..(((((...(((((((---------.....)))))))..((..((((....((((((...(((((((.....))))))).)))))).))))..)))))))(((((........))))). ( -35.10, z-score = -2.43, R) >droYak2.chr2R 2627796 111 + 21139217 UGGUCAUGCCCAUGGUG---------CACAACGCCAUGCCCACCUUUGAUGAUUUUGACUUUGAGGAGUUCCUCUCCUCAUUCGAGAACGAGAAUGACGACGAGCAAUUCCCGCUGUUCA ..(((((...(((((((---------.....)))))))...........((.((((((...(((((((.....))))))).)))))).))...)))))...(((((........))))). ( -33.40, z-score = -2.14, R) >droSec1.super_1 10464356 111 - 14215200 UGGUCAUGCCCAUGGUG---------CACAACGCCAUGCCCACCUUCGAUGAUUUUGACUUCGAGGAGUUCCUCUCCUCAUUCGAGAACGAGAAUGACGACGAGCAAUUCCCGCUGUUCA ..(((((...(((((((---------.....)))))))......(((((.((........))((((((.....))))))..))))).......)))))...(((((........))))). ( -32.30, z-score = -1.39, R) >droSim1.chr2R 11692212 111 - 19596830 UGGUCAUGCCCAUGGUG---------CACAACGCCAUGCCCACCUUCGAUGAUUUUGACUUCGAGGAGUUCCUCUCCUCAUUCGAGAACGAGAAUGACGACGAGCAAUUCCCGCUGUUCA ..(((((...(((((((---------.....)))))))......(((((.((........))((((((.....))))))..))))).......)))))...(((((........))))). ( -32.30, z-score = -1.39, R) >consensus UGGUCAUGCCCAUGGUG_________CA_AACGCCAUGCCCACCUUCGAUGAUUUUGACUUUGAGGAGUUCCUCUCCUCAUUCGAGAACGAGAAUGACGACGAGCAAUUCCCGCUGUUCA ........................................................(((..((.((((((.(((((.((((((........)))))).)).))).))))))))..))).. (-18.15 = -17.50 + -0.65)

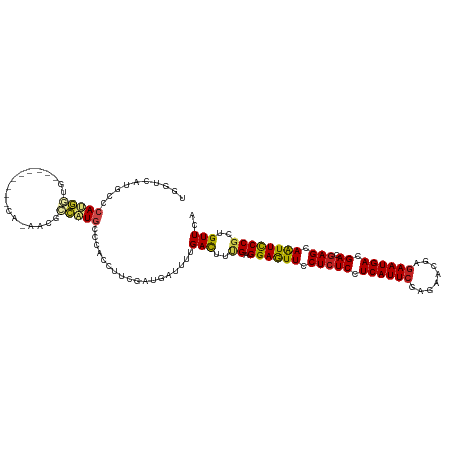
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:52 2011