| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,913,905 – 12,914,055 |
| Length | 150 |
| Max. P | 0.897023 |

| Location | 12,913,905 – 12,914,055 |
|---|---|
| Length | 150 |
| Sequences | 10 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 65.39 |
| Shannon entropy | 0.70298 |
| G+C content | 0.46881 |
| Mean single sequence MFE | -45.26 |
| Consensus MFE | -13.98 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12913905 150 + 21146708 --AC-CAAUUCUGGAGGAGGCCAGUUCAUGACGCCAGAAGUGCGUCCCUAGGCGGCGCACGAAACAGG------------AAAGUCCAAACUACAACAUGGCAUUUAUAGGAUUACUGCUCUACUACCUGAUGGUUUUGCUCUUCUACCUCCUGGGUCCGGAUUU--- --..-....(((((((((((..(((((((((.((((...((((((((....).)))))))......((------------.....))...........))))..)))).)))))...((......(((....)))...)).......)))))....))))))...--- ( -41.80, z-score = 0.20, R) >droSim1.chr2R 11650004 150 + 19596830 --AC-CAAUUCUGGAGGAGACCAGUUCAUGACGCCAGAAGUGCGCCCCUUGGCGGCGCACGAAACAGG------------AAAGUCCAAACUACAACAUGGCAUUUAUAGGAUUACUGCUCUACUACCUGAUGGUAUUGCUCUUUUACCUCCUGGGUCCGGAUUU--- --.(-(.((((.((((((((.((((((..((.((((...((((((((....).)))))))......((------------.....))...........)))).)).....))..)))).)))..((((....))))...........))))).))))..))....--- ( -45.50, z-score = -1.37, R) >droSec1.super_1 10422376 150 + 14215200 --AC-CAAUUCUGGAGGAGACCAGUUCAUGACGCCAGAAGUGCGCCCCUUGGCGGCGCACGAAACAGG------------AAAGUCCAAACUACAACAUGGCAUUUAUAGGAUUACUGCUCUACUACCUGAUGGUAUUGCUCUUUUACCUCUUGGGUCCGGAUUU--- --..-....((((((((((...(((((((((.((((...((((((((....).)))))))......((------------.....))...........))))..)))).)))))....))))....((..(.((((.........))))..)..))))))))...--- ( -45.00, z-score = -1.35, R) >droYak2.chr2R 2585708 150 - 21139217 --AC-CAAUUUUGGAGGAAACCAGCUCAUGACGCCAGAAGUGCGCCCCUUGGCGGCGCACAAAACAUG------------AAAGUCCAAACUACAACAUGGCAUUUUUAGGAUUACUGCUUUACUACCUGGUGGUAUUGCUAUUCUACCUCCUGGGUCCAGAUUU--- --.(-((.....(((((......((.((((.........((((((((....).)))))))........------------..(((....)))....)))))).....((((((....((..((((((...))))))..)).))))))))))))))..........--- ( -44.10, z-score = -1.63, R) >droEre2.scaffold_4845 9661011 150 - 22589142 --AC-CAAUUUUGGAGGAGGCCAGCUCAUGACGCCAGAAGUACGCCCCUUGGCGGCGCACGAUACAGG------------AAAGUCCAAACUACAACAUGGCAUUUUUAGGAUUAGUGCUCUACUACCUGGUGGUAUUGCUAUUCUACCUCCUGGGUCCAGAUUU--- --.(-((.....((((((((..(((......((((((.((((((((....))))..((((....(..(------------((((((((..........))).))))))..)....))))..))))..)))))).....)))..))).))))))))..........--- ( -44.20, z-score = -0.37, R) >droAna3.scaffold_13266 6198281 153 - 19884421 AGACACAAUACUCGAGGAGGAGACCUCAUGACGCCAGUGGUGCGCCUCUCUGGGGCGCACACACCCGG------------AUAGCGAACCCUACAACAUGGCAUUUAUAGGCUUACUUAUCUAUUACCUAGUGGUAUUGUUAUUCUACCUCCUGGGACCCGAUUU--- ..........(((.((((((................(((((((((((.....)))))))).)))..((------------(((((((((((((....((((.....(((((....)))))))))....))).))).)))))))))..))))))))).........--- ( -48.10, z-score = -1.82, R) >dp4.chr3 12452098 153 - 19779522 ---GCUGGUUGUCGACGAAUGCAGCUCAUGACGCCAGUCGUGCGCCACUUCCGGGCGCACGGGUGAGG------------GCAAUGGACAUUUCCAGUAUGCAUUUAUAGGAUUAAUUCUCUCUUACUUAGUCGCAAUGAUAUUUUACUGCCUCAACCAGCACUAUGC ---((((((((..((((((((((((((....((((...((((((((.......)))))))))))).))------------))..((((....))))...))))))).(((((.........)))))....)))(((.(((....))).)))..))))))))....... ( -55.80, z-score = -3.49, R) >droPer1.super_2 1508035 153 - 9036312 ---GCUGGUUGUCGACGAAUGCAGCUCAUGACGCCAGUCGUGCGCCACUUCCGGGCGCACGGGUGAGG------------GCAAUGGACAUUUCCAGUAUGCAUUUAUAGGAUUAAUUCUCUCUUACUUAGUCGCAAUGAUAUUUUACUGCCUCAACCAGCACUAUGC ---((((((((..((((((((((((((....((((...((((((((.......)))))))))))).))------------))..((((....))))...))))))).(((((.........)))))....)))(((.(((....))).)))..))))))))....... ( -55.80, z-score = -3.49, R) >droWil1.scaffold_180700 4933669 162 + 6630534 ---GCAGAUUGUCGAUCAGUCUGACAGUUGACGCCAGAAUCUUGCCAAGUAUUGGCGCAAAAAUCACACACAUGCCAGCAGGCAUACUCCCUGUCGUUACGCAUUUAUAGGAUUACUUCUAUAUUAUUUUUUAGUAUUGCUUUUUCACUGUCUUCACGUCCAAUU--- ---..((((((.....))))))(((((((((((.(((......((((.....))))...............(((((....))))).....))).))))).(((..(((((((....))))))).((((....)))).)))......)))))).............--- ( -36.80, z-score = -1.51, R) >droVir3.scaffold_13324 430528 125 - 2960039 ----------------------------UGACGCCGAUUGUGUGCCUUAUUUUGGCACACACAAAACG------------GUUAUCGACUUUACCAAAUCGCACUAAUAGGAUUAUUACUUUAUUAUCUAGUGAUAUUAGUGUUUUAUUGCGUCAAUUUGCUUUU--- ----------------------------((((((.(((((((((((.......))))))).......(------------((..........))).))))(((((((((..((((.............))))..)))))))))......))))))..........--- ( -35.52, z-score = -4.53, R) >consensus __AC_CAAUUCUGGAGGAGACCAGCUCAUGACGCCAGAAGUGCGCCCCUUGGCGGCGCACGAAACAGG____________AAAGUCCAAACUACAACAUGGCAUUUAUAGGAUUACUGCUCUACUACCUGGUGGUAUUGCUAUUUUACCUCCUGGGUCCGGAUUU___ .......................................(((((((.......)))))))....((((.......................................((((...............))))..((((.........)))).)))).............. (-13.98 = -13.40 + -0.58)

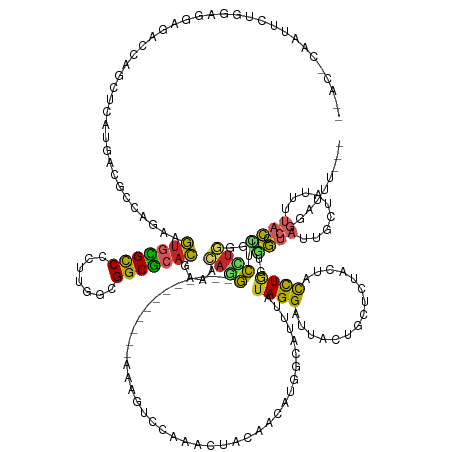

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:43 2011