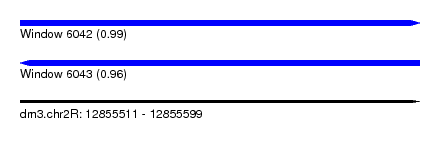
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,855,511 – 12,855,599 |
| Length | 88 |
| Max. P | 0.986397 |

| Location | 12,855,511 – 12,855,599 |
|---|---|
| Length | 88 |
| Sequences | 13 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Shannon entropy | 0.39721 |
| G+C content | 0.57266 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -30.35 |
| Energy contribution | -30.24 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
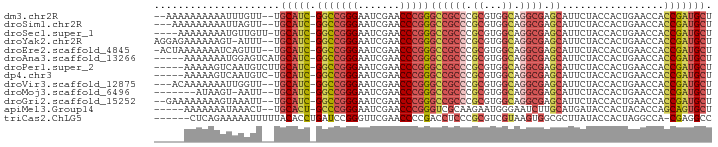
>dm3.chr2R 12855511 88 + 21146708 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA--AACAAAUUUUUUUUUU-- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).--................-- ( -35.90, z-score = -2.08, R) >droSim1.chr2R 11591930 87 + 19596830 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA--AACUAAUUUUUUUUU--- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).--...............--- ( -35.90, z-score = -2.09, R) >droSec1.super_1 10364102 87 + 14215200 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA-AACAACAUUUUUUUU---- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).-...............---- ( -35.90, z-score = -2.01, R) >droYak2.chr2R 2526925 89 - 21139217 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA--AAAU-ACUUUUUUCUCCU .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).--....-............. ( -35.90, z-score = -1.79, R) >droEre2.scaffold_4845 11190845 89 + 22589142 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA--AAACUGAUUUUUUUAGU- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).--..(((((.....)))))- ( -37.80, z-score = -2.25, R) >droAna3.scaffold_13266 6142402 87 - 19884421 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCAUGACUCCAUUUUUUU----- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-)))))................----- ( -35.90, z-score = -1.49, R) >droPer1.super_2 7026725 87 + 9036312 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCAAGACAUUGACUUUUU----- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-)))))................----- ( -35.90, z-score = -1.50, R) >dp4.chr3 6819457 86 + 19779522 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA-GACAUUGACUUUUU----- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).-..............----- ( -35.90, z-score = -1.53, R) >droVir3.scaffold_12875 10888531 87 + 20611582 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA--AACCAAUUUUUUUGU--- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).--...............--- ( -35.90, z-score = -1.59, R) >droMoj3.scaffold_6496 10426577 82 - 26866924 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA--AAUU-ACUUAU------- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).--....-......------- ( -35.90, z-score = -2.04, R) >droGri2.scaffold_15252 11469470 88 + 17193109 AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC-GAUGCA--AAUUUACUUUUUUUUC-- .(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))-))))).--................-- ( -35.90, z-score = -2.28, R) >apiMel3.Group14 2180900 85 + 8318479 AGCACUGCUGGUGUAGUGGUAUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGC-AGUGCA--AGUUUAUUUUUUU----- .((((((((((((......)))))(((((((.......)))))))..(((((.......)))))))-))))).--.............----- ( -32.20, z-score = -3.70, R) >triCas2.ChLG5 14941700 86 + 18847211 GGCCUCG-UGGCCUAGUGGUAUAAGCGCCACUUACGACGCGGGAGGUCGGGGUUCGAACCCGGAUCAGGUGUAAAAAUUUUUCUGAG------ ((((...-.)))).((((((......))))))..((((.(....)))))((((....))))...(((((............))))).------ ( -29.20, z-score = -0.79, R) >consensus AGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCC_GAUGCA__AAAUUAUUUUUUU_____ .((((((((.((((.(((((((.........)))))))..)))).)))((((.......))))....)))))..................... (-30.35 = -30.24 + -0.12)
| Location | 12,855,511 – 12,855,599 |
|---|---|
| Length | 88 |
| Sequences | 13 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 82.72 |
| Shannon entropy | 0.39721 |
| G+C content | 0.57266 |
| Mean single sequence MFE | -31.31 |
| Consensus MFE | -23.09 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
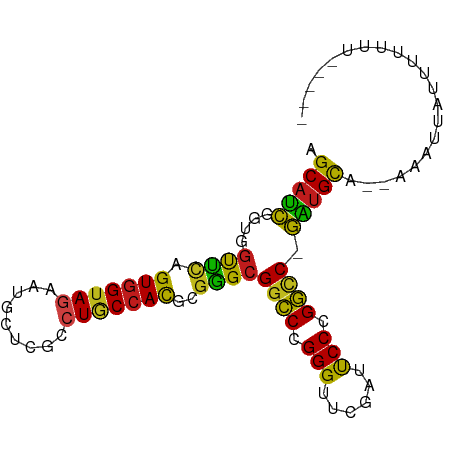
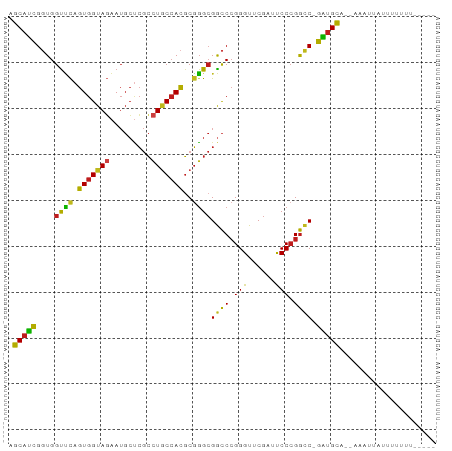
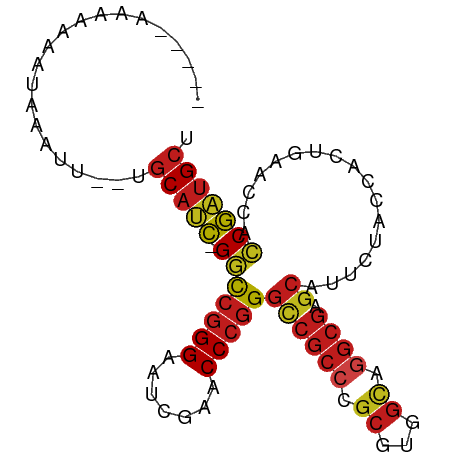
>dm3.chr2R 12855511 88 - 21146708 --AAAAAAAAAAUUUGUU--UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU --................--.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.91, R) >droSim1.chr2R 11591930 87 - 19596830 ---AAAAAAAAAUUAGUU--UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU ---...............--.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -2.08, R) >droSec1.super_1 10364102 87 - 14215200 ----AAAAAAAAUGUUGUU-UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU ----...............-.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.61, R) >droYak2.chr2R 2526925 89 + 21139217 AGGAGAAAAAAGU-AUUU--UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU .............-....--.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.37, R) >droEre2.scaffold_4845 11190845 89 - 22589142 -ACUAAAAAAAUCAGUUU--UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU -.................--.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.94, R) >droAna3.scaffold_13266 6142402 87 + 19884421 -----AAAAAAAUGGAGUCAUGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU -----................(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.34, R) >droPer1.super_2 7026725 87 - 9036312 -----AAAAAGUCAAUGUCUUGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU -----................(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.48, R) >dp4.chr3 6819457 86 - 19779522 -----AAAAAGUCAAUGUC-UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU -----..............-.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.56, R) >droVir3.scaffold_12875 10888531 87 - 20611582 ---ACAAAAAAAUUGGUU--UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU ---...............--.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.46, R) >droMoj3.scaffold_6496 10426577 82 + 26866924 -------AUAAGU-AAUU--UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU -------......-....--.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -1.99, R) >droGri2.scaffold_15252 11469470 88 - 17193109 --GAAAAAAAAGUAAAUU--UGCAUC-GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU --................--.(((((-(((((((.......)))))((((((.((...)).)))).)).................))))))). ( -32.20, z-score = -2.20, R) >apiMel3.Group14 2180900 85 - 8318479 -----AAAAAAAUAAACU--UGCACU-GCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAUACCACUACACCAGCAGUGCU -----.............--.(((((-(((((((.......)))))...((((((.......)))))).................))))))). ( -29.30, z-score = -3.89, R) >triCas2.ChLG5 14941700 86 - 18847211 ------CUCAGAAAAAUUUUUACACCUGAUCCGGGUUCGAACCCCGACCUCCCGCGUCGUAAGUGGCGCUUAUACCACUAGGCCA-CGAGGCC ------(((..................(((.((((.(((.....)))...)))).)))....(((((.((.........))))))-))))... ( -23.50, z-score = -1.02, R) >consensus _____AAAAAAAUAAAUU__UGCAUC_GGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCU .....................(((((.(((((((.......)))))((((((.((...)).)))).)).................))))))). (-23.09 = -23.64 + 0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:38 2011