
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,850,525 – 12,850,644 |
| Length | 119 |
| Max. P | 0.845465 |

| Location | 12,850,525 – 12,850,617 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.78 |
| Shannon entropy | 0.50328 |
| G+C content | 0.35289 |
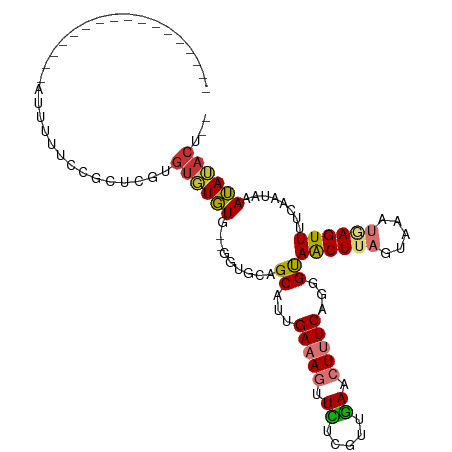
| Mean single sequence MFE | -20.21 |
| Consensus MFE | -9.63 |
| Energy contribution | -9.60 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
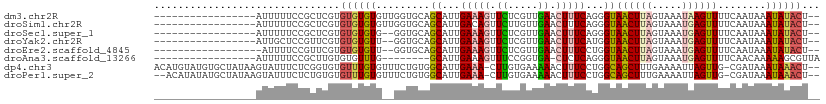
>dm3.chr2R 12850525 92 + 21146708 -----------------AUUUUUCCGCUCGUGUGUGUGUUGGUGCAGCAUUGAAAGUUCUCGUUGAACUUUCAGGGUAACUUAGUAAAUAAGUUUUCAAUAAAUAUACU-- -----------------............((((((.((((((....((.((((((((((.....)))))))))).))((((((.....)))))).)))))).)))))).-- ( -24.90, z-score = -3.52, R) >droSim1.chr2R 11586750 92 + 19596830 -----------------AUUUUUCCGCUCGUGUGUGUGUUGGUGCAGCAUUGACAGUUCUUGUUGAACUUUCAGGGUAACUUAGUAAAUGAGUUUUCAAUAAAUAUACU-- -----------------............((((((.((((((....((.((((.(((((.....))))).)))).))((((((.....)))))).)))))).)))))).-- ( -21.50, z-score = -1.44, R) >droSec1.super_1 10353897 90 + 14215200 -----------------AUUUUUCCGCUCGUGUGUGUG--GGUGCAGCAUUGAAAGUUCUCGUUGAACUUUCAGGGUAACUUAGUAAAUGAGUUUUCAAUAAAUAUACU-- -----------------........((((((.....((--(((...((.((((((((((.....)))))))))).)).)))))....))))))................-- ( -21.60, z-score = -1.83, R) >droYak2.chr2R 2521205 90 - 21139217 -----------------AUUGCUCCGUUCGUGUGUGUU--GGUGCAGCAUUGAAAGUUCUCGUUGAACUUUCAUGGUAACUUAGUAAAUGAGUUUUCAAUAAAUAUACU-- -----------------.((((.(((...........)--)).))))...(((((((((.....)))))))))(((.((((((.....)))))).)))...........-- ( -22.00, z-score = -2.32, R) >droEre2.scaffold_4845 9595558 89 - 22589142 ------------------AUUUUCCGUUCGUGUGUGUU--GGUGCAGCAUUGAAAGUUCUCGUUGAACUUUCCUGGUAACUUAGUAAAUGAGUUUUCAAUAAAUAUACU-- ------------------...........(((((((((--(...))))...((((((((.....)))))))).(((.((((((.....)))))).)))....)))))).-- ( -19.50, z-score = -2.08, R) >droAna3.scaffold_13266 6137928 85 - 19884421 -----------------AUUUUUCCGCUUGUGUGUUUG--------GCAUUGAAAGUUUCCGGUGA-CUCUCAGGGUAACUUAGUAAAUGAGUUUUCAACAAAAAGCGUUA -----------------.......(((((...((((..--------...((((.((((......))-)).))))((.((((((.....)))))).))))))..)))))... ( -18.20, z-score = -0.72, R) >dp4.chr3 6814189 107 + 19779522 ACAUGUAUGUGCUAUAAGUAUUUCUCGGUGUGUUUGUGUUUCUGUGGCAUUGAAA-CUUGUGAAAAACUUUCCUGGCAGCUUUGAAAAUUAGUUG-CGAUAAAUAAACU-- ....((.(((..(((((((.((((.(((.((....(((((.....)))))....)-)))).)))).)))).....((((((.(....)..)))))-).))).))).)).-- ( -17.40, z-score = 0.68, R) >droPer1.super_2 7021455 105 + 9036312 --ACAUAUAUGCUAUAAGUAUUUCUCUGUGUGUUUGUGUUUCUGUGGCAUUGAAA-CUUGUGAAAAACUUUCCUGGCAGCUUUGAAAAUUAGUUG-CGAUAAAUAAACU-- --(((((.((((.....)))).....)))))(((((((((((.........))))-)..................((((((.(....)..)))))-).....)))))).-- ( -16.60, z-score = 0.25, R) >consensus _________________AUUUUUCCGCUCGUGUGUGUG__GGUGCAGCAUUGAAAGUUCUCGUUGAACUUUCAGGGUAACUUAGUAAAUGAGUUUUCAAUAAAUAUACU__ ...............................((((((.........((...(((((.((.....)).)))))...))((((((.....))))))........))))))... ( -9.63 = -9.60 + -0.03)
| Location | 12,850,537 – 12,850,644 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.59 |
| Shannon entropy | 0.51973 |
| G+C content | 0.37050 |
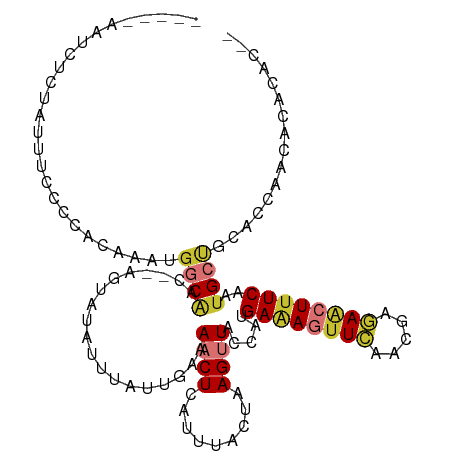
| Mean single sequence MFE | -17.96 |
| Consensus MFE | -7.20 |
| Energy contribution | -7.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12850537 107 - 21146708 -----AAUCUCUAUUUUCCCACAAAUGGCAAC--AGUAUAUUUAUUGAAAACUUAUUUACUAAGUUACCCUGAAAGUUCAACGAGAACUUUCAAUGCUGCACCAACACACACAC -----....................(((...(--(((((..........((((((.....))))))....(((((((((.....)))))))))))))))..))).......... ( -21.20, z-score = -4.13, R) >droSim1.chr2R 11586762 107 - 19596830 -----AAUCUCUAUUUCCCCACAAAUGGCAAC--AGUAUAUUUAUUGAAAACUCAUUUACUAAGUUACCCUGAAAGUUCAACAAGAACUGUCAAUGCUGCACCAACACACACAC -----....................(((...(--((((((((((.((((......)))).))))).....(((.(((((.....))))).)))))))))..))).......... ( -15.50, z-score = -1.43, R) >droSec1.super_1 10353909 105 - 14215200 -----AAUCUCUAUUUCCCCACAAAUGGCAAC--AGUAUAUUUAUUGAAAACUCAUUUACUAAGUUACCCUGAAAGUUCAACGAGAACUUUCAAUGCUGCACCCACACACAC-- -----....................(((...(--((((((((((.((((......)))).))))).....(((((((((.....)))))))))))))))...))).......-- ( -19.00, z-score = -3.30, R) >droYak2.chr2R 2521217 104 + 21139217 -----AAUCUUC-UUUCCCCACAAAUGGCAAC--AGUAUAUUUAUUGAAAACUCAUUUACUAAGUUACCAUGAAAGUUCAACGAGAACUUUCAAUGCUGCACCAACACACAC-- -----.......-............(((...(--((((((((((.((((......)))).))))).....(((((((((.....)))))))))))))))..)))........-- ( -19.20, z-score = -3.24, R) >droEre2.scaffold_4845 9595569 104 + 22589142 -----AAUCUCU-UUUCCCCACAAAUGGCAAC--AGUAUAUUUAUUGAAAACUCAUUUACUAAGUUACCAGGAAAGUUCAACGAGAACUUUCAAUGCUGCACCAACACACAC-- -----.......-............(((...(--(((((.....(((..((((.........))))..)))((((((((.....)))))))).))))))..)))........-- ( -18.30, z-score = -2.47, R) >droAna3.scaffold_13266 6137940 104 + 19884421 UAUCCCAACGUUAUCCCUUC-CAAAUGGCUACUAACGCUUUUUGUUGAAAACUCAUUUACUAAGUUACCCUGAGAG-UCACCGGAAACUUUCAAUGCCAAACACAC-------- ....................-....((((.......((((.....(((....)))......)))).....((((((-(........)))))))..)))).......-------- ( -13.40, z-score = -0.27, R) >dp4.chr3 6814220 103 - 19779522 -------ACUGUGCAUUCAAAUACUCCUCGGUGCAGUUUAUUUAUCG-CAACUAAUUUUCAAAGCUGCCAGGAAAGUUUUUCACAAG-UUUCAAUGCCACAGAAACACAAAC-- -------.(((((((((.((((..((((..(.(((((((........-.............))))))))))))..((.....))..)-))).)))).)))))..........-- ( -19.00, z-score = -1.18, R) >droPer1.super_2 7021484 103 - 9036312 -------ACUGUGCAUUCAAAUACUCGUCGGUGCAGUUUAUUUAUCG-CAACUAAUUUUCAAAGCUGCCAGGAAAGUUUUUCACAAG-UUUCAAUGCCACAGAAACACAAAC-- -------.(((((((((.(((((((..((.(.(((((((........-.............)))))))).))..))).........)-))).)))).)))))..........-- ( -18.10, z-score = -0.59, R) >consensus _____AAUCUCUAUUUCCCCACAAAUGGCAAC__AGUAUAUUUAUUGAAAACUCAUUUACUAAGUUACCAUGAAAGUUCAACGAGAACUUUCAAUGCUGCACCAACACACAC__ ..........................((((...................((((.........)))).....((((((((.....))))))))..))))................ ( -7.20 = -7.70 + 0.50)

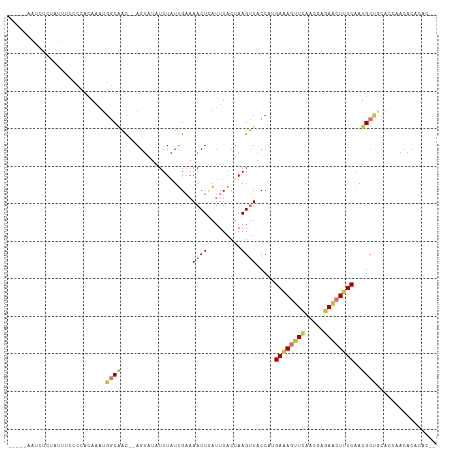
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:36 2011