| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,819,979 – 12,820,034 |
| Length | 55 |
| Max. P | 0.989932 |

| Location | 12,819,979 – 12,820,034 |
|---|---|
| Length | 55 |
| Sequences | 13 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Shannon entropy | 0.42997 |
| G+C content | 0.46896 |
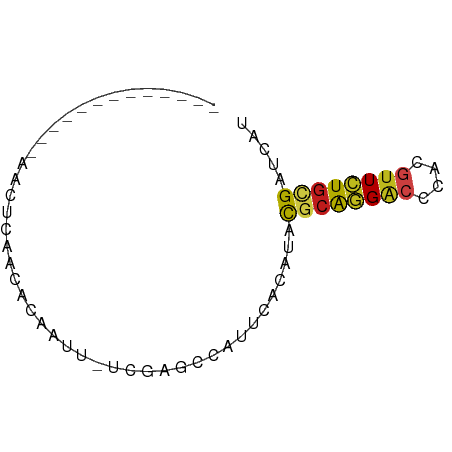
| Mean single sequence MFE | -10.86 |
| Consensus MFE | -8.19 |
| Energy contribution | -7.99 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12819979 55 + 21146708 -------------AACUCAACACAAUU-UCGAGCCAUUCACAUACGCAGGACCCACGUUCUGCGAUCAU -------------..(((.........-..)))...........((((((((....))))))))..... ( -12.20, z-score = -3.51, R) >droSim1.chr2R 11556287 55 + 19596830 -------------AACUCAACACAAUU-UCGAGCCAUUCACAUACGCAGGACCCACGUUCUGCGAUCAU -------------..(((.........-..)))...........((((((((....))))))))..... ( -12.20, z-score = -3.51, R) >droSec1.super_1 10323582 55 + 14215200 -------------AACUCAACACAAUU-UCGAGCCAUUCACAUACGCAGGACCCACGUUCUGCGAUCAU -------------..(((.........-..)))...........((((((((....))))))))..... ( -12.20, z-score = -3.51, R) >droYak2.chr2R 2489622 55 - 21139217 -------------AACUCAACACAAUU-UCGAGCCAUUCACAUACGCAGGACCCACGUUCUGCGAUCAU -------------..(((.........-..)))...........((((((((....))))))))..... ( -12.20, z-score = -3.51, R) >droEre2.scaffold_4845 9563425 55 - 22589142 -------------AACUCAACACAAUU-UCGAGCCAUUCACAUACGCAGGACCCACCUUCUGCGAUCAU -------------..(((.........-..)))...........(((((((......)))))))..... ( -9.50, z-score = -3.02, R) >droAna3.scaffold_13266 6105512 55 - 19884421 -------------AACUCAACACAAUU-UCGAGCCAUUCACAUAUGCAGGACCCACGUUCUGCGAUCAU -------------..(((.........-..)))...........((((((((....))))))))..... ( -10.20, z-score = -2.12, R) >dp4.chr2 17631862 68 - 30794189 GAUUCAACGUCAAUGUGCCCCACCGGU-UUGUGGUGCACAGCUACAAGCGGUUCACCUUCUGCGACCAC ........(((..(((((.((((....-..)))).))))).......((((........)))))))... ( -20.70, z-score = -1.25, R) >droPer1.super_2 6989296 56 + 9036312 -------------AACUCAACACAAUAAUCGAACCAUUCACAUAUGCAGGACCCACGUUCUGUGACCAU -------------................................(((((((....)))))))...... ( -7.40, z-score = -0.85, R) >droWil1.scaffold_180700 6565918 55 + 6630534 -------------AACUCAACAUCACU-UCGAACCAUUCACAUAUGCAGGACCCACGUUCUGUGAUCAU -------------..............-.................(((((((....)))))))...... ( -7.40, z-score = -0.60, R) >droVir3.scaffold_12875 18089707 55 - 20611582 -------------AACUCAGCACCAUU-UCGAACCAUUUACAUACGCAGGACCCACGUUCUGUGACCAU -------------..............-................((((((((....))))))))..... ( -10.00, z-score = -2.34, R) >droMoj3.scaffold_6496 22718904 55 + 26866924 -------------AACUCAGCACCAUU-UCGAACCAUUCACAUAUGCAGGACCCACGUUCUGUGACCAU -------------..............-.................(((((((....)))))))...... ( -7.40, z-score = -0.53, R) >droGri2.scaffold_15112 529042 55 - 5172618 -------------AACUCAGCACCAUU-UCGAACCAUUCACAUACGCAGGACCCACGUUCUGUGAUCAU -------------..............-................((((((((....))))))))..... ( -10.00, z-score = -1.96, R) >anoGam1.chr3L 38689106 55 + 41284009 -------------UACCAAGCAUGAGU-UUAAAGAGUGGACCUAUUCGAGUCCAACGUUUUGCGAUCAC -------------.........(((..-.(((((..(((((........)))))...)))))...))). ( -9.80, z-score = -0.72, R) >consensus _____________AACUCAACACAAUU_UCGAGCCAUUCACAUACGCAGGACCCACGUUCUGCGAUCAU ............................................((((((((....))))))))..... ( -8.19 = -7.99 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:33 2011