| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,814,459 – 12,814,578 |
| Length | 119 |
| Max. P | 0.990810 |

| Location | 12,814,459 – 12,814,578 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.63 |
| Shannon entropy | 0.60295 |
| G+C content | 0.37630 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -13.16 |
| Energy contribution | -15.28 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
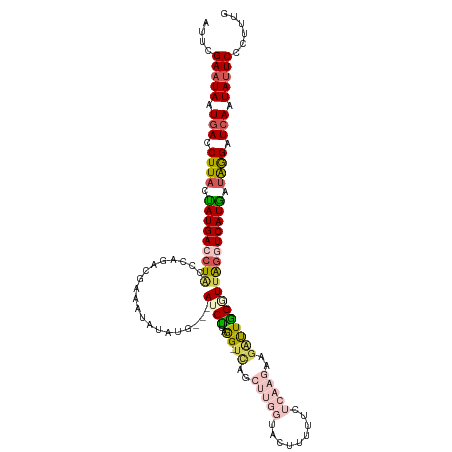
>dm3.chr2R 12814459 119 + 21146708 CAAAGGGAAUAUUGAUCCUAUCAUGACCUGACCUAAUCUACUUGAGAAAAAGUACCAAGCUGG-CUUAAGAUAUACAUAUAUUUCGUUUGGGUAGGUCAUAGUAAGGUCACUAUUCGAAU ......(((((.(((((.(((.((((((((.(((((.(..((((((....(((.....)))..-))))))((((....))))...).))))))))))))).))).))))).))))).... ( -34.50, z-score = -3.28, R) >droSim1.chr2R 11550987 119 + 19596830 -CAAGGGAAUAUUGAUCCUAUCAUGACUUAAACCAAUCCACUUGAGAAAAAGUACCAAGCUGAUCCUAGGAUCCUAUCCUAUUUCGUAUGGGUAGCUCAUAUUAAGAUCAUUAUUCGAAU -.....(((((.(((((.((..((((.(((.........((((......)))).(((.((.((...((((((...))))))..)))).))).))).))))..)).))))).))))).... ( -26.10, z-score = -1.22, R) >droSec1.super_1 10318237 119 + 14215200 -UAAGGGAUUAUUGAUCCUAUCAUGACCUAAACCAAUCCUCUUGAGAAAAAGUACCAAGCUGAUCCUAGGAUCCUAUCCUAUUUCGUAUGGGUAGCUCAUAUUAAGAUCAUUAUUCUAAU -...(((((...(((((.((..((((.(((..........(((......)))..(((.((.((...((((((...))))))..)))).))).))).))))..)).)))))..)))))... ( -24.20, z-score = -0.41, R) >droYak2.chr2R 2484027 99 - 21139217 CAAAUGGAAUACUGAUCUUAUUAUGACCUAACCUAAUCUUAUUAAG---------------AA-CUUAAGAU---CAUAUGCUCA--CAUGGCAGGUCAUAGAAAGGUCAUUAUUCGGUA ...((.(((((.(((((((.(((((((((......((((((.....---------------..-..))))))---.....(((..--...)))))))))))).))))))).))))).)). ( -30.60, z-score = -4.86, R) >droEre2.scaffold_4845 9558316 91 - 22589142 CAAAUGGAAUAUUGAUCUCAUCAUGACCCAAUCCAGCU-----------------------GA-CCCGAGAU---CAUAUAGCCG--CCGGGCAGGUCAUGGUAAGGUGAUUAUUCGGAA ......(((((.(.((((.(((((((((.........(-----------------------(.-((((....---..........--.))))))))))))))).)))).).))))).... ( -25.96, z-score = -0.82, R) >consensus CAAAGGGAAUAUUGAUCCUAUCAUGACCUAAACCAAUCCACUUGAGAAAAAGUACCAAGCUGA_CCUAAGAU___CAUAUAUUUCGUAUGGGUAGGUCAUAGUAAGGUCAUUAUUCGAAU ......(((((.(((((.(((.((((((((.(((.......................................................))))))))))).))).))))).))))).... (-13.16 = -15.28 + 2.12)


| Location | 12,814,459 – 12,814,578 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.63 |
| Shannon entropy | 0.60295 |
| G+C content | 0.37630 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -13.33 |
| Energy contribution | -13.69 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12814459 119 - 21146708 AUUCGAAUAGUGACCUUACUAUGACCUACCCAAACGAAAUAUAUGUAUAUCUUAAG-CCAGCUUGGUACUUUUUCUCAAGUAGAUUAGGUCAGGUCAUGAUAGGAUCAAUAUUCCCUUUG ....(((((.(((.((((.((((((((..((....................(((((-....)))))(((((......))))).....))..)))))))).)))).))).)))))...... ( -27.90, z-score = -2.03, R) >droSim1.chr2R 11550987 119 - 19596830 AUUCGAAUAAUGAUCUUAAUAUGAGCUACCCAUACGAAAUAGGAUAGGAUCCUAGGAUCAGCUUGGUACUUUUUCUCAAGUGGAUUGGUUUAAGUCAUGAUAGGAUCAAUAUUCCCUUG- ....(((((.((((((((.(((((.(((((.........((((((...)))))).((((.((((((.........)))))).)))))))...))))))).)))))))).))))).....- ( -31.90, z-score = -1.80, R) >droSec1.super_1 10318237 119 - 14215200 AUUAGAAUAAUGAUCUUAAUAUGAGCUACCCAUACGAAAUAGGAUAGGAUCCUAGGAUCAGCUUGGUACUUUUUCUCAAGAGGAUUGGUUUAGGUCAUGAUAGGAUCAAUAAUCCCUUA- ....((.((.((((((((.(((((.(((.((........((((((...)))))).((((..(((((.........)))))..))))))..))).))))).)))))))).)).)).....- ( -31.20, z-score = -1.23, R) >droYak2.chr2R 2484027 99 + 21139217 UACCGAAUAAUGACCUUUCUAUGACCUGCCAUG--UGAGCAUAUG---AUCUUAAG-UU---------------CUUAAUAAGAUUAGGUUAGGUCAUAAUAAGAUCAGUAUUCCAUUUG ....(((((.(((.(((..((((((((((((((--....))).((---((((((..-..---------------.....)))))))))).)))))))))..))).))).)))))...... ( -29.40, z-score = -4.60, R) >droEre2.scaffold_4845 9558316 91 + 22589142 UUCCGAAUAAUCACCUUACCAUGACCUGCCCGG--CGGCUAUAUG---AUCUCGGG-UC-----------------------AGCUGGAUUGGGUCAUGAUGAGAUCAAUAUUCCAUUUG ....(((((.....((((.((((((((..((((--(((((.....---......))-))-----------------------.)))))...)))))))).)))).....)))))...... ( -28.10, z-score = -1.57, R) >consensus AUUCGAAUAAUGACCUUACUAUGACCUACCCAGACGAAAUAUAUG___AUCUUAGG_UCAGCUUGGUACUUUUUCUCAAGAAGAUUGGGUUAGGUCAUGAUAGGAUCAAUAUUCCCUUUG ....(((((.(((.((((.(((((((((..............................................................))))))))).)))).))).)))))...... (-13.33 = -13.69 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:32 2011