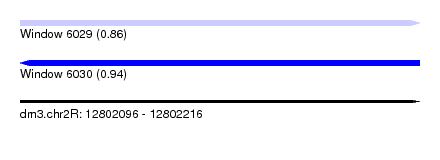
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,802,096 – 12,802,216 |
| Length | 120 |
| Max. P | 0.940320 |

| Location | 12,802,096 – 12,802,216 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.64 |
| Shannon entropy | 0.07821 |
| G+C content | 0.47653 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -32.52 |
| Energy contribution | -32.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12802096 120 + 21146708 GCCAUUCGUCCUUGUAGGGCUUGCAUCUGCUACAUCUUCAAACAUAUUAAACAUCAGCCACUAUAACAUCAACAGGUUCCCCCGUUGAUGAUGGCUGACCUAAUGAUGUUUCAGCGGGGC (((....((((.....))))......((((((((((.........((((....(((((((......(((((((.((....)).))))))).)))))))..)))))))))...)))))))) ( -35.40, z-score = -2.28, R) >droSim1.chr2R 11538411 119 + 19596830 GCCAUUCGUCCUUGUAGGGCUUGCAUCUGCUACAUCUUCGAGCAUAUUAAACAUCAGCCACUACAACAUCAACAGGUUCCCC-GUUGAUGAUGGCUGACCUAAUGAUGUUCCAGCGGGGC (((....((((.....))))......(((((........((((((((((....(((((((......(((((((.((....))-))))))).)))))))..)))).)))))).)))))))) ( -36.20, z-score = -1.76, R) >droSec1.super_1 10306139 119 + 14215200 GCCAUUUGUCCUUGUAGGGCUUGCAUCUGCUACAUCUUCAAACAUAUUAAACAUCAGCCACUACAACAUCAACAGGUUCCCC-GUUGAUGAUGGCUGACCUAAUGAUGUUCCAGCGGGGC (((....((((.....))))......((((((((((.........((((....(((((((......(((((((.((....))-))))))).)))))))..)))))))))...)))))))) ( -34.10, z-score = -1.71, R) >droYak2.chr2R 2470985 119 - 21139217 GCCAUUCGUUCUUGUAGAGCUUGCAUCUGCUACAUCUUCAGACAUAUUAGACAUCAGCCACUACAACAUCAACAGGUUCCCC-GUGGAUGAUGGCUGACCUAAUGAUGUUCCAGCGGGGC (((....((((.....))))......((((((((((.........(((((...(((((((......((((.((.((....))-)).)))).))))))).))))))))))...)))))))) ( -31.60, z-score = -0.45, R) >droEre2.scaffold_4845 9546212 119 - 22589142 GCCAUUCGUCCUUGUAGGGCUUGCAUCUGCUACAUCUUCAAACAUAUUAAACAUCAACCACUACAACAUCAACAGGUUCCCC-GUUGAUGAUGGCUGACCUAAUGAUGUUCCAGCGGGGU (((....((((.....))))..((....))...........................((.((..(((((((..(((((..((-(((...)))))..)))))..)))))))..)).))))) ( -29.00, z-score = -0.64, R) >consensus GCCAUUCGUCCUUGUAGGGCUUGCAUCUGCUACAUCUUCAAACAUAUUAAACAUCAGCCACUACAACAUCAACAGGUUCCCC_GUUGAUGAUGGCUGACCUAAUGAUGUUCCAGCGGGGC (((....((((.....))))......((((((((((.........((((....(((((((......(((((((.((....)).))))))).)))))))..)))))))))...)))))))) (-32.52 = -32.44 + -0.08)

| Location | 12,802,096 – 12,802,216 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Shannon entropy | 0.07821 |
| G+C content | 0.47653 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -36.54 |
| Energy contribution | -36.38 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12802096 120 - 21146708 GCCCCGCUGAAACAUCAUUAGGUCAGCCAUCAUCAACGGGGGAACCUGUUGAUGUUAUAGUGGCUGAUGUUUAAUAUGUUUGAAGAUGUAGCAGAUGCAAGCCCUACAAGGACGAAUGGC (((...(((..(((((.....(((((((((((((((((((....)))))))))).....))))))))).(((((.....))))))))))..)))......(.((.....)).)....))) ( -44.00, z-score = -3.72, R) >droSim1.chr2R 11538411 119 - 19596830 GCCCCGCUGGAACAUCAUUAGGUCAGCCAUCAUCAAC-GGGGAACCUGUUGAUGUUGUAGUGGCUGAUGUUUAAUAUGCUCGAAGAUGUAGCAGAUGCAAGCCCUACAAGGACGAAUGGC (((..(((....((((.....((((((((((((((((-(((...)))))))))).....)))))))))........(((((......).))))))))..)))((.....))......))) ( -39.30, z-score = -1.70, R) >droSec1.super_1 10306139 119 - 14215200 GCCCCGCUGGAACAUCAUUAGGUCAGCCAUCAUCAAC-GGGGAACCUGUUGAUGUUGUAGUGGCUGAUGUUUAAUAUGUUUGAAGAUGUAGCAGAUGCAAGCCCUACAAGGACAAAUGGC (((..(((...(((((.....((((((((((((((((-(((...)))))))))).....))))))))).(((((.....)))))))))).((....)).)))((.....))......))) ( -39.60, z-score = -2.12, R) >droYak2.chr2R 2470985 119 + 21139217 GCCCCGCUGGAACAUCAUUAGGUCAGCCAUCAUCCAC-GGGGAACCUGUUGAUGUUGUAGUGGCUGAUGUCUAAUAUGUCUGAAGAUGUAGCAGAUGCAAGCUCUACAAGAACGAAUGGC (((.(((((.(((((((.(((((...((.((......-)))).))))).))))))).)))))(((..(((((..((((((....))))))..)))))..)))...............))) ( -38.10, z-score = -1.45, R) >droEre2.scaffold_4845 9546212 119 + 22589142 ACCCCGCUGGAACAUCAUUAGGUCAGCCAUCAUCAAC-GGGGAACCUGUUGAUGUUGUAGUGGUUGAUGUUUAAUAUGUUUGAAGAUGUAGCAGAUGCAAGCCCUACAAGGACGAAUGGC ...(((((...(((((.....((((((((((((((((-(((...)))))))))).....))))))))).(((((.....)))))))))).((....)).)))((.....))......)). ( -35.40, z-score = -1.29, R) >consensus GCCCCGCUGGAACAUCAUUAGGUCAGCCAUCAUCAAC_GGGGAACCUGUUGAUGUUGUAGUGGCUGAUGUUUAAUAUGUUUGAAGAUGUAGCAGAUGCAAGCCCUACAAGGACGAAUGGC (((..(((.((((((.(((((((((((((((((((((.((....)).))))))).....)))))))))..))))))))))).....((((.....)))))))((.....))......))) (-36.54 = -36.38 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:27 2011