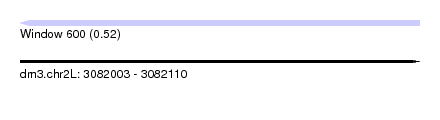
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,082,003 – 3,082,110 |
| Length | 107 |
| Max. P | 0.522500 |

| Location | 3,082,003 – 3,082,110 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 57.52 |
| Shannon entropy | 0.80738 |
| G+C content | 0.43386 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -6.05 |
| Energy contribution | -5.90 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3082003 107 - 23011544 -UCUGGCUAUAAAUUUAUGCGGUAACAUAAAACCGUCAACGGUUGAUUAUGGCCACACAUUGGAUAUACAUUUGUGU------AUAUCCAUGCCGUAAAGUUUUGGGCUUGGCC -((.((((..((((((.(((((((.((((((((((....)))))..))))).........((((((((((....)))------))))))))))))))))))))..)))).)).. ( -36.50, z-score = -3.45, R) >droVir3.scaffold_12963 19976252 88 - 20206255 -CUUAGCCAUAAAUUGAUGCGGUAACAUGG---CCAAAGUCAAUGAGUAUGGCGCGUGCUUGUAAGGAAAUUGUGGAA--AUUACGGCUAGGCA-------------------- -....((((((.((((((..(((......)---))...))))))...))))))((.((((.((((.............--.))))))).).)).-------------------- ( -24.64, z-score = -1.14, R) >droSim1.chr2L 3020133 107 - 22036055 -UCUGGCUAUGAAUUUAUGCGGUAACAUAAAACCGUCAACGCUUGAUUAUGGCCACACAUCGGAUAUACAUUUGUGU------AUAUCCAUGCCGCAAAGUUUUGGGCUUGGCC -..(((((((((..(((.(((...((........))...))).))))))))))))......(((((((((....)))------))))))..(((.(((....))))))...... ( -32.00, z-score = -1.99, R) >droSec1.super_5 1234058 106 - 5866729 -UCUGGCUAUAAAUUUAUGCGGUAACAUAAAACCGUCAACGCUUGAUUAUGGCCACACAUCGGAUAUACAUUUGUGU------AUAUCCAUG-CGCAAAGUUUUGGGCUUGGCC -..(((((((((..(((.(((...((........))...))).))))))))))))..(((.(((((((((....)))------)))))))))-.((.((((.....)))).)). ( -31.10, z-score = -2.16, R) >droEre2.scaffold_4929 3124513 83 - 26641161 -UCUGGCUAUAAAUUUAUGCGGUAACAUAAAACCGUCAACGGUCGAUUAUGGCCACACAUCGGAUAUACAUUUGGGU------UUUGGCC------------------------ -..(((((((((.((((((......))))))((((....))))...))))))))).....(((((....)))))(((------....)))------------------------ ( -19.20, z-score = -0.80, R) >droYak2.chr2L 3072877 112 - 22324452 -UCUGGCUAUAAAUUUAUGCGGUAACAUAAAACCGUCAACGGUUGAUUAUGGCCGCACAUCGGAUAUAUGUACAUAUACAUCCAUAUCCAUGCCGUAAAG-UUUGGGUUUGGCC -...((((((((((((.(((((((.((((((((((....)))))..)))))..........(((((((((((....)))))..)))))).))))))))))-))))....))))) ( -33.80, z-score = -2.12, R) >droAna3.scaffold_12913 298285 94 - 441482 -UUUGGCCAUAAAUUUAUACGGUAACAUAAAACCGUCAGCCGUUGAUUAUGGCCUCCUUGGAUUUGGUUGCUCUUUUCCAUUUUUUCUUUUGGCC------------------- -...(((((.........(((((........)))))..(((((.....))))).....((((...((.....))..))))..........)))))------------------- ( -21.70, z-score = -1.34, R) >dp4.chr4_group1 670988 88 + 5278887 UCGUAGUCGGAAAUUUAUGUGGUAACAUAAAACCGUCAACCGUUGUCUAUGGCAGACACAUGGCCGUAUGCUCGUAGACCUGGCCGGC-------------------------- .....(((((...(((((((....)))))))........(((..((((((((((.((........)).))).))))))).))))))))-------------------------- ( -26.30, z-score = -0.98, R) >droPer1.super_5 603953 88 - 6813705 UCUUAUUCGGAAAUUUAUGUGGUAACAUAAAACCGUCAACCGUCGUCUAUGGCAGACACAUGGCCGUAUGCUCGUAGACCAGGCCGGC-------------------------- ......((((...(((((((....)))))))........((...((((((((((.((........)).))).)))))))..)))))).-------------------------- ( -24.20, z-score = -0.99, R) >consensus _UCUGGCUAUAAAUUUAUGCGGUAACAUAAAACCGUCAACGGUUGAUUAUGGCCACACAUCGGAUAUACAUUCGUGU______AUAGCCAUG_C____________________ .....((((((.......(((((........)))))...........))))))............................................................. ( -6.05 = -5.90 + -0.14)

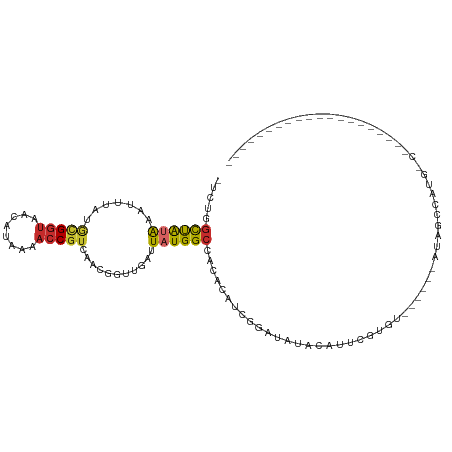
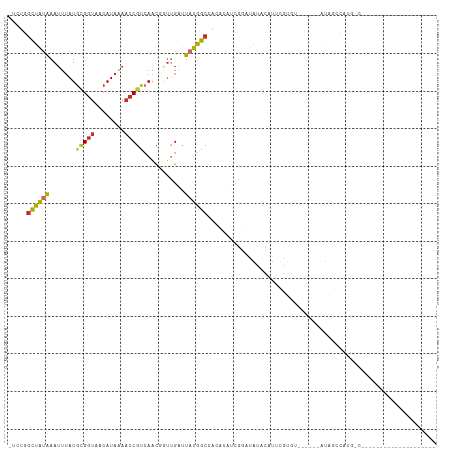
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:59 2011