
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,777,244 – 12,777,340 |
| Length | 96 |
| Max. P | 0.500000 |

| Location | 12,777,244 – 12,777,340 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 64.90 |
| Shannon entropy | 0.69254 |
| G+C content | 0.47458 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -9.81 |
| Energy contribution | -9.21 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 12777244 96 + 21146708 CUUGUCUUCUGUGCACGUUUGUGUGUA---CA--GACUAUAUAGUAUAAUGGUGCAGACAUCGCGUGACCUGCGCUAAUGCAUG--GCCGCGCCCCAAGAGAC ..(((((((((((((((....))))))---))--)).......((((....)))))))))..(((((.((((((....)))).)--).))))).......... ( -32.60, z-score = -1.81, R) >droSim1.chr2R 11512870 96 + 19596830 CUUGUAGUCUAUGCACGUUUGUGUGUA---CA--GACUAUAUAGUAUAAUGGUGCAGACAUCGCGUGACCUGCGCUAAUGCAUG--GCCGCGCCCCAAGAGAC ..((((((((.((((((....))))))---.)--))))))).........((((....))))(((((.((((((....)))).)--).))))).......... ( -30.70, z-score = -1.20, R) >droSec1.super_1 10281450 96 + 14215200 CUUGUCUUCUAUGCACGUUUGUGUGUA---CA--GACUAUAUAGUAUAAUGGUGCAGACAUCGCGUGACCUGCGCUAAUGCAUG--GCCGCGCCCCAAGAGAC ..((((((((.((((((....))))))---.)--)).......((((....)))))))))..(((((.((((((....)))).)--).))))).......... ( -26.60, z-score = -0.19, R) >droYak2.chr2R 4941701 98 - 21139217 CUUGUCUUUUAUGCACGUUUGUGUGUG---UACCAACUAUAUAGUAUAAUGGUGCAGACAUCGCGUGACCUGCGCUAAUGCAUG--GCCGCGCCCCAAGAGAC ...(((((((..........((((.((---((((((((....)))....))))))).)))).(((((.((((((....)))).)--).)))))...))))))) ( -32.50, z-score = -2.13, R) >droAna3.scaffold_13266 6063410 86 - 19884421 --------CCAGUCUCGAACGAGUAUG---UA----GUAUAUAGUAUAAUGGUGCAGACAUCGCGUGACUUGCGCUAAUGCACU--GGCGCACCCCGAGGGGG --------..((((.((..(((((.((---((----.(((........))).)))).)).))))).))))(((((((......)--))))))(((....))). ( -28.50, z-score = -1.06, R) >droWil1.scaffold_180700 6469312 74 + 6630534 -----------------GACACACACA---CAC---UUACAUAUACUUAUAGAGUAGACAUCGCGUGAGUUGCCUUAAUGCAAGU-GCCACACCCCAA----- -----------------(.(((.....---.((---((((...(((((...)))))(....)..))))))(((......))).))-).).........----- ( -10.10, z-score = -0.15, R) >droVir3.scaffold_12875 18029262 79 - 20611582 -------UAUAUACACUAUUGUAUAUA---CA-----UGCAGGAUUUAAUAGCGCAGACAUCGCGUGAGCUGCGCUAAUGCAUU--GCCACACCCU------- -------((((((((....))))))))---.(-----((((........((((((((...((....)).)))))))).))))).--..........------- ( -21.40, z-score = -1.64, R) >droGri2.scaffold_15112 460822 95 - 5172618 CUAUUAUUUUGAUCGCAACUGUGAGUAACGCACACUAUAUGCAAUACAAUGGCUCAGACAUCGCGUGAGUUGCGCUAAUGCAUAUUGCCACACCC-------- ..............((((.((((..((.((((.(((.(((((......(((.......))).)))))))))))).))...)))))))).......-------- ( -17.00, z-score = 1.32, R) >droMoj3.scaffold_6496 22648738 79 + 26866924 -------UUUAUAGUUUAGUGUAUACA---UA-----UAUACGCUUUAAUAGCGCAGACAUCGCGUGAGCUGCGCUAAUGCAUU--GCCACACCCU------- -------..........((((((((..---..-----))))))))....((((((((...((....)).)))))))).......--..........------- ( -20.30, z-score = -1.27, R) >consensus CU__U__UCUAUGCACGAUUGUGUGUA___CA____CUAUAUAGUAUAAUGGUGCAGACAUCGCGUGACCUGCGCUAAUGCAUG__GCCGCACCCCAAG_G__ .................................................((((((((...((....)).)))))))).......................... ( -9.81 = -9.21 + -0.60)
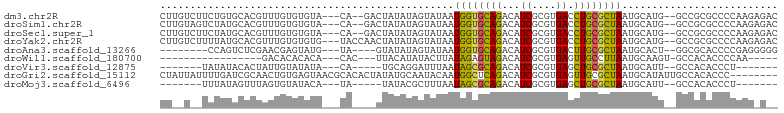
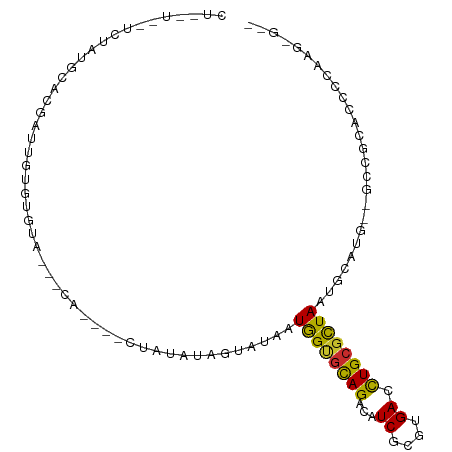

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:23 2011