
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,757,484 – 12,757,577 |
| Length | 93 |
| Max. P | 0.630272 |

| Location | 12,757,484 – 12,757,577 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.35 |
| Shannon entropy | 0.68247 |
| G+C content | 0.34335 |
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -7.11 |
| Energy contribution | -7.32 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 12757484 93 - 21146708 ---------ACUUAAUA-CCAAUUAAUUUC---CGC-CAAUGUAAUAGAUG-UAUACUAAUCGGAAGUAAUUUCAAUGGCAAUGAUGCGUUAUUGAAAACUAACGAUG--- ---------........-.......(((((---((.-....(((.......-..)))....)))))))..((((((((((........))))))))))..........--- ( -15.50, z-score = -1.00, R) >droPer1.super_2 6926076 103 - 9036312 ACAAGACAUUUUUCAAG-CGAAUUAAUUUC---CGUCUAAUGUAAGGGGGGCUACACAAAUCGGAAGCGAUUUCAAUGGCAAUGCCGUGUUAUUGAAACCA--CCGCAG-- ................(-((......((((---((.....((((........)))).....)))))).(.(((((((((((......))))))))))).).--.)))..-- ( -22.60, z-score = -0.40, R) >dp4.chr3 6703005 103 - 19779522 ACAAGACAUUUUUCAAG-CGAAUUAAUUUC---CGUCUAAUGUAAGGGGGGCUACACAAAUCGGAAGCGAUUUCAAUGGCAAUGCCGUGUUAUUGAAACCA--CCGCAG-- ................(-((......((((---((.....((((........)))).....)))))).(.(((((((((((......))))))))))).).--.)))..-- ( -22.60, z-score = -0.40, R) >droAna3.scaffold_13266 6042751 99 + 19884421 -------AUGUUUAAUGUUGAAAAAGUUUC---CGCGUAAUGUAAUGGGA--UAUACAAAUCGGAAGUAAUUUCAAUGGCAUUAUUGUGUUAUUGAAAAAAACCGGCAAAA -------........(((((......((((---((.....((((......--..))))...))))))...((((((((((((....)))))))))))).....)))))... ( -21.70, z-score = -1.93, R) >droEre2.scaffold_4845 9494481 93 + 22589142 ---------ACUUAAUA-CCAAUUAAUUUC---CGC-AAAUGUAAUGAAUG-UAUACGAAUCGGAAGUAAUUUCAAUGGCAAUGAUGAGUUAUUGAAAACUAAC-GCAG-- ---------........-.......(((((---((.-...((((.......-..))))...)))))))..((((((((((........))))))))))......-....-- ( -15.70, z-score = -1.00, R) >droYak2.chr2R 4920771 93 + 21139217 ---------ACUUAAUA-CCAAUUAAUUUC---CGC-CAAUGUAAUAGAUG-UACACUAAUCGGAAGUAAUUUCAAUGGCAAUGAUGCGUUAUUGAAAACUA-ACGCAG-- ---------........-............---.((-((........(((.-.......)))((((....))))..)))).....(((((((.(....).))-))))).-- ( -19.20, z-score = -2.28, R) >droSec1.super_1 10261887 89 - 14215200 ---------ACUUAAUA-CGAAUUAAUUUC---CGC-CAAUGUA----AUG-UAUACUAAUCGGAAGUAAUUUCAAUGGCAAUGAUGCGUUAUUGAAAACUAACGAUG--- ---------........-((.....(((((---((.-....(((----...-..)))....)))))))..((((((((((........)))))))))).....))...--- ( -17.30, z-score = -1.82, R) >droSim1.chr2R 11492828 93 - 19596830 ---------ACUUAAUA-CCAAUUAAUUUC---CGC-CAAUGUAAUGGAUG-UAUACUAAUCGGAAGUAAUUUCAAUGGCAAUGAUGCGUUAUUGAAAACUAACGAUG--- ---------........-............---..(-((......)))...-.......((((..(((..((((((((((........)))))))))))))..)))).--- ( -16.60, z-score = -1.01, R) >droMoj3.scaffold_6496 22619572 92 - 26866924 -----------UUCAAAUUAAUUCCGUUUCAUUAGCUCAAUUUAAAUAAUA-UGUUAAUGUCGGAAGUAGUU--AGCGAUGCCA--AUGUAAUUGAAAACCUCCUCUG--- -----------(((((((((.(((((...(((((((...............-.))))))).))))).)))))--.....(((..--..)))..))))...........--- ( -15.09, z-score = -1.86, R) >droGri2.scaffold_15112 432722 90 + 5172618 --------CGCAGGCAAUAAAUUUAGUUCC----GCUUUAUGUAACAAGUC-UAUUAAUAUCGGAAGUAUUU--GGCCACAAUGACACGUAACUGAAAUCAUCUG------ --------..((((..((...(((((((.(----(..((((.......(((-....((((.(....))))).--)))....))))..)).)))))))))..))))------ ( -11.20, z-score = 1.04, R) >consensus _________ACUUAAUA_CCAAUUAAUUUC___CGC_CAAUGUAAUGGAUG_UAUACAAAUCGGAAGUAAUUUCAAUGGCAAUGAUGCGUUAUUGAAAACUAACGGCA___ .............................................................(....)...((((((((((........))))))))))............. ( -7.11 = -7.32 + 0.21)

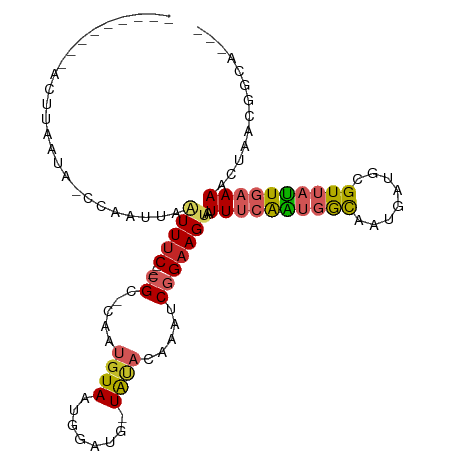
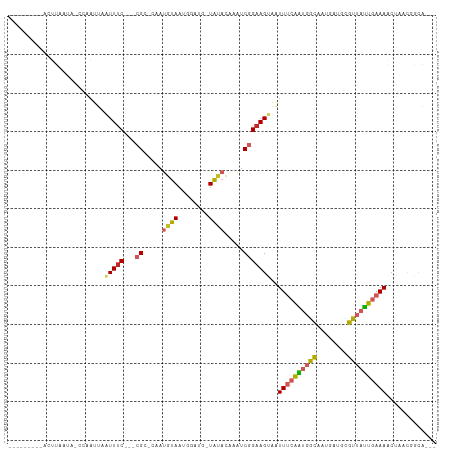
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:21 2011