| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,744,912 – 12,745,008 |
| Length | 96 |
| Max. P | 0.779774 |

| Location | 12,744,912 – 12,745,008 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.31 |
| Shannon entropy | 0.58095 |
| G+C content | 0.36960 |
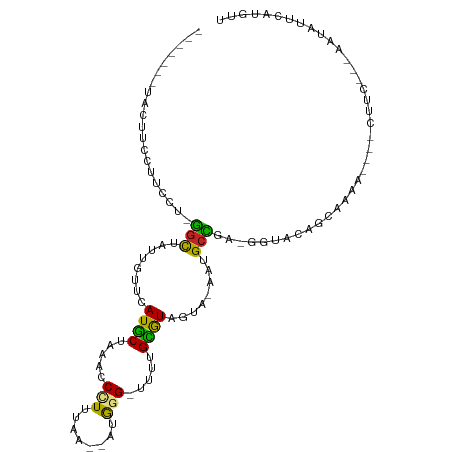
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -7.33 |
| Energy contribution | -6.75 |
| Covariance contribution | -0.58 |
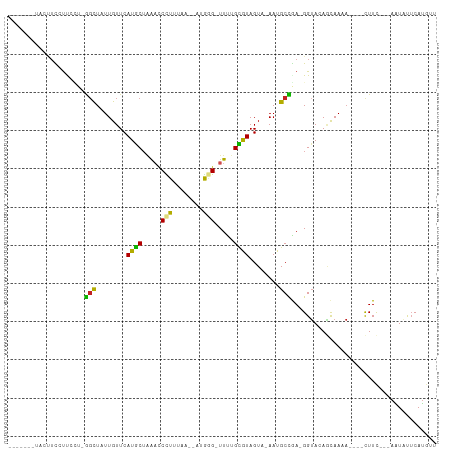
| Combinations/Pair | 1.75 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
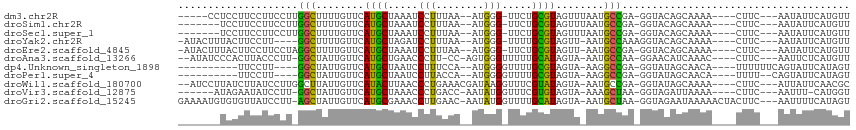
>dm3.chr2R 12744912 96 + 21146708 -----CCUCCUUCCUUCCUUGGCUUUUGUUCAUGCUAAAUCCUUUAA--AUGGG-UUCUGCGUAGUUUAAUGCCGA-GGUACAGCAAAA----CUUC---AAUAUUCAUGUU -----...........(((((((..(((..(((((..(((((.....--..)))-))..)))).)..))).)))))-))..........----....---............ ( -22.40, z-score = -2.56, R) >droSim1.chr2R 11480203 94 + 19596830 -------UCCUUCCUUCCUUGGCUUUUGUUCAUGCUAAAUCCUUUAA--AUGGG-UUCUGCGUAGUUUAAUGCCGA-GGUACAGCAAAA----CUUC---AAUAUUCAUGUU -------.........(((((((..(((..(((((..(((((.....--..)))-))..)))).)..))).)))))-))..........----....---............ ( -22.40, z-score = -2.65, R) >droSec1.super_1 10249301 94 + 14215200 -------UCCUUCCUUCCUUGGCUUUUGUUCAUGCUAAAUCCUUUAA--AUGGG-UUCUGCGUAGUUUAAUGCCGA-GGUACAGCAAAA----CUUC---AAUAUUCAUGUU -------.........(((((((..(((..(((((..(((((.....--..)))-))..)))).)..))).)))))-))..........----....---............ ( -22.40, z-score = -2.65, R) >droYak2.chr2R 4908110 96 - 21139217 -AUACUUUACUUCCUU----GGCUUUUGUUCAUGCUAGAUCCUUUAA--AUGGG-UUUUGCGUAGUU-AAUGCCAAAGGUACAGCAAAA----CUUC---AAUAUUCAUGUU -...((.(((((..((----(((..(((...((((.((((((.....--..)))-))).))))...)-)).)))))))))).)).....----....---............ ( -19.90, z-score = -1.25, R) >droEre2.scaffold_4845 9482013 99 - 22589142 -AUACUUUACUUCCUUCCUAGGCUUUUGUUCAUGCUAAAUCCUUUAA--AUGGG-UUCUGCGUAGUU-AAUGCCGA-GGUACAGCAAAA----CUUC---AAUAUUCAUGUU -...................(((..(((...((((..(((((.....--..)))-))..))))...)-)).)))((-(((........)----))))---............ ( -18.70, z-score = -1.00, R) >droAna3.scaffold_13266 6030244 98 - 19884421 --AUAUCCCACUUACCCUU-GGCUAUUGUUCAUGCUGAACCCUU-CC-AGUGGGUUUUUGCAUAGUA-AAUGCCAA-GGAACAUCAAAC----CUUC---AAUUCUCAUGUU --.............((((-(((.(((....((((.((((((..-..-...))))))..))))....-))))))))-))..........----....---............ ( -25.90, z-score = -3.07, R) >dp4.Unknown_singleton_1898 55636 90 - 80150 ----------UUCCUU----GGCUAUUGUUCAUGCUAAUCCUUUCCA--AUGGGGUUUUGCGUAGUA-AAGGCCGA-GGUAUAGCAACA----UUUUUUCAGUAUUCAUAGU ----------..((((----((((.(((((.((((.((((((.....--..))))))..))))))))-).))))))-))..........----................... ( -22.90, z-score = -1.99, R) >droPer1.super_4 6568924 88 + 7162766 ----------UUCCUU----GGCUAUUGUUCAUGCUAAUCCUUACCA--AUGGGGUUUUGCGUAGUA-AAGGCCGA-GGUAUAGCAACA----UUUU--CAGUAUUCAUAGU ----------..((((----((((.(((((.((((.((((((.....--..))))))..))))))))-).))))))-))..........----....--............. ( -22.90, z-score = -1.71, R) >droWil1.scaffold_180700 6420105 101 + 6630534 --AUCCUUAUCUUAUCCUUGGCUUAUUGUUCAUACUUAACCCUGAAACGAUAAGGUUUCGUAUAGUA-AAUGCCGA-GGUAUAGCAAAA----CUUC---AUUAUUCAACGC --.........((((((((((((((((((.............((((((......)))))).))))))-)..)))))-)).)))).....----....---............ ( -17.24, z-score = -0.84, R) >droVir3.scaffold_12875 9968769 94 - 20611582 ------AUAGAAUAUCCUU-GGCUAUUGUUCAUGCUAAACCCUGACC-AAUAUGGUUUCGUGUAGUA-AAAGCUAA-GGUAGAUUAAAA----CUUC---AAUUU-CAUGGU ------....(((..((((-((((.(((((((((..(((((......-.....))))))))).))))-).))))))-))...)))....----....---.....-...... ( -17.50, z-score = -0.52, R) >droGri2.scaffold_15245 14569736 105 - 18325388 GAAAAUGUGUGUUAUCCUU-AGCUAUUGUUCAUGCGAAACCUUGAAC-AAUAUGGUUUUGCAUAGUA-AAUGCUAA-GGUAGAAUAAAAACUACUUC---AAUUUUCAUAGU (((...((.((((..((((-(((.(((....((((((((((......-.....))))))))))....-))))))))-))...))))...))...)))---............ ( -28.20, z-score = -4.01, R) >consensus _______UACUUCCUUCCU_GGCUAUUGUUCAUGCUAAACCCUUUAA__AUGGG_UUUUGCGUAGUA_AAUGCCGA_GGUACAGCAAAA____CUUC___AAUAUUCAUGUU ....................(((........((((..((.(((........))).))..))))........)))...................................... ( -7.33 = -6.75 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:20 2011