
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,728,021 – 12,728,182 |
| Length | 161 |
| Max. P | 0.822283 |

| Location | 12,728,021 – 12,728,182 |
|---|---|
| Length | 161 |
| Sequences | 5 |
| Columns | 171 |
| Reading direction | reverse |
| Mean pairwise identity | 78.19 |
| Shannon entropy | 0.37171 |
| G+C content | 0.46672 |
| Mean single sequence MFE | -51.86 |
| Consensus MFE | -31.46 |
| Energy contribution | -34.58 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
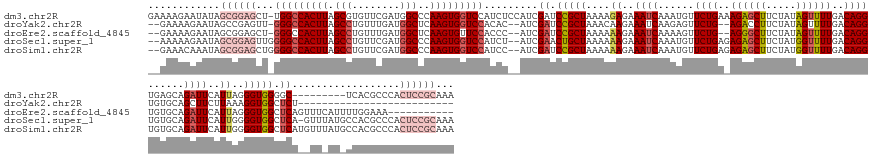
>dm3.chr2R 12728021 161 - 21146708 GAAAAGAAUAUAGCGGAGCU-UGGCCACUUAGCGUGUUCGAUGGCCCAAGUGGUCCAUCUCCAUCGAUCCGCUAAAAGAGAAAUCAAAUGUUCUGAAAGAGCUUCUAUAGUUUUGACAGGUGAGCAGAUUCAUUAGGGUGGGGC---------UCACGCCCACUCCGCAAA ............((((((..-.(((...((((((.(.(((((((.((....)).......))))))).)))))))..((....)).......(((..((((((.....))))))..)))((((((..((((....))))...))---------)))))))..))))))... ( -52.40, z-score = -1.87, R) >droYak2.chr2R 4891220 138 + 21139217 --GAAAAGAAUAGCCGAGUU-GGGCCACUUAGCCUGUUUGAUGGCUCAAGUGGUCCACAC--AUCGAUCCGCUAAACAAGAAAUCAAGAGUUCUG--AGACCUUCUAUAGUUUUGACAGGUGUGCAGCUUCUUAAAGGUGGCUCU-------------------------- --.............(((((-(((((((((((((........)))).))))))))))..(--((((((...((.....))..)))(((((..(((--(.((((((.........)).)))).).))).)))))...)))))))).-------------------------- ( -40.90, z-score = -1.04, R) >droEre2.scaffold_4845 9467062 153 + 22589142 --GAAAAGAAUAGCGGAGCU-GGGCCACUUAGCCUGUUUGAUGGCUCAAGUGUUCCACCC--AUCGAUCCGCUAAAAAAGAAAUCAAAAGUUCUG--AGGGCUUCUAUAGUUUUGACAGGUGUGCAGAUUCAUUAGGGUGGCUCAGUUUCAUUUUGGAAA----------- --............((((((-(((((((((((((........))))....(((..((((.--.((((...((((.(.(((...(((.......))--)...))).).)))).))))..)))).))).........)))))))))))))))..........----------- ( -44.50, z-score = -0.75, R) >droSec1.super_1 10232582 166 - 14215200 --AAAAAGAAUAGCGGAGUUGGGGCCACUUAGCCUGUUCGAUGGCCCAAGUGGUCCAUCU--AUCGAACUGCUAAAAAAGAAAUCAAAUGUUCUGAGAGAGCUUCUAUGGUUUUGACAGGUGUGCAGAUUCAUUGGGGUGGCUCA-GUUUAUGCCACGCCCACUCCGCAAA --..........(((((((..(((((((((.(((........)))..)))))))))....--...(((((((.......(((((((...((((.....)))).....))))))).........)))).)))...((((((((...-......))))).))))))))))... ( -61.99, z-score = -3.40, R) >droSim1.chr2R 11463382 167 - 19596830 --GAAACAAAUAGCGGAGCUGGGGCCACUUAGCCUGUUCGAUGGCCCAAGUGGUCCAUCC--AUCGAUCCGCUAAAAAAGAAAUCAAAUGUUCUGAGAGAGCUUCUAUGGUUUUGACAGGUGUGCAGAUUCAUUGGGGUGGCUCAUGUUUAUGCCACGCCCACUCCGCAAA --..........((((((((((((((((((.(((........)))..)))))))))..((--((.((...(((.....((((........)))).....))).)).))))......)))..............(((((((((..........))))).))))))))))... ( -59.50, z-score = -2.35, R) >consensus __GAAAAGAAUAGCGGAGCU_GGGCCACUUAGCCUGUUCGAUGGCCCAAGUGGUCCAUCC__AUCGAUCCGCUAAAAAAGAAAUCAAAUGUUCUGA_AGAGCUUCUAUAGUUUUGACAGGUGUGCAGAUUCAUUAGGGUGGCUCA__UU_AU_CCACGCCCACUCCGCAAA ............((((((...(((((((((.(((........)))..))))))))).........((.(((((....((..((((......((((..((((((.....))))))..))))......))))..))..))))).))..................))))))... (-31.46 = -34.58 + 3.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:15 2011