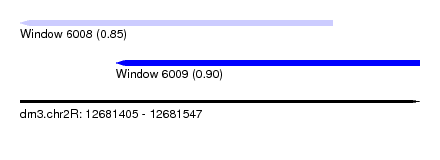
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 12,681,405 – 12,681,547 |
| Length | 142 |
| Max. P | 0.904405 |

| Location | 12,681,405 – 12,681,516 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.31 |
| Shannon entropy | 0.49321 |
| G+C content | 0.38507 |
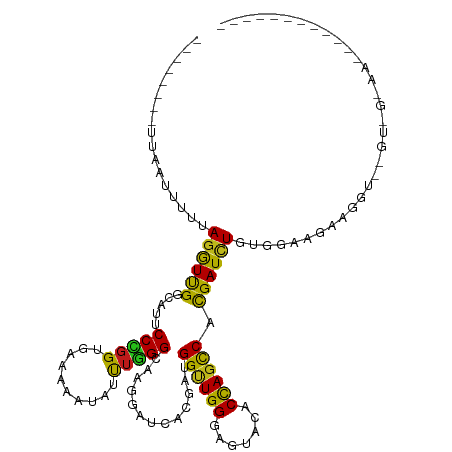
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -15.32 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
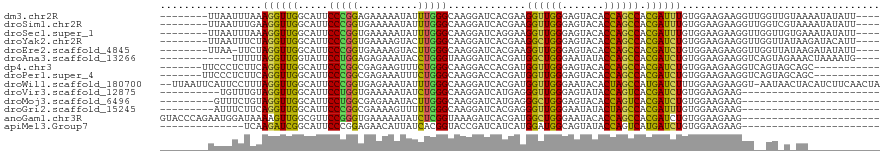
>dm3.chr2R 12681405 111 - 21146708 AUAUUUGGGCAAGGAUCACGAAGGUUGGGAGUACACCAGCCACGAUUUGUGGAAGAAGGUUGGUUGUAAAAUAUAUUUUAAGUUUAAGGAUGUUAUUUAAACACUUAAAUC- ((((((..((((..........((((((.......)))))).((((((........)))))).)))).)))))).......(((((((..((((.....))))))))))).- ( -22.20, z-score = -1.04, R) >droSec1.super_1 10186199 111 - 14215200 AUAUUUGGGCAAGGAUCAGGAAGGUUGGGAGUACACCAGCCACGAUUUGUGGAAGAAGGUUGGUUGUGAAAUAUAUUUUAAGUUUAAUUAUGUUAUUUAAUCACAUAAACC- .........((..((((.(...((((((.......)))))).)))))..))......((((...(((((((((((.((.......)).))))))......)))))..))))- ( -21.60, z-score = -0.47, R) >droYak2.chr2R 4843174 111 + 21139217 GUACUUGGGCAAGGAUCACGAAGGCUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGUUGGUUAUAAGAUACAUUUGAAGUAUAAGAAUGCCCUUUAAUCACUUAAAUC- ......(((((...........((((((.......)))))).((((((........))))))........((((.......)))).....)))))................- ( -27.60, z-score = -1.08, R) >droEre2.scaffold_4845 9418714 111 + 22589142 GUACUUGGGCAAGGAUCACGAAGGUUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGUUGGUUAUAAGAUAUAUUUGAAGUUUAGGGAUGCCAUUUAAUCACCUAAAUU- .........((((.(((.....((((((.......)))))).((((((........)))))).......)))...)))).(((((((((((........))).))))))))- ( -28.40, z-score = -1.30, R) >droAna3.scaffold_13266 5970170 105 + 19884421 AUACCUGGGUAAGGAUCACGAUGGCUGGGAAUAUACCAGCCACGAUCUGUGGAAGAAGGUCAGUAGAAACUAAAAUGCCAGAGUUCGUAAUCU------AUAACUUAUAUU- .....(((((..((((.(((((((((((.......))))))).(((((........))))).......................)))).))))------...)))))....- ( -27.20, z-score = -1.48, R) >droWil1.scaffold_180700 3751629 111 + 6630534 AUAUUUGGGCAAGGAUCACGAUGGUUGGGAAUACACUAGCCAUGAUCUUUGGAAGAAGGU-AAUAACUACAUCUUCAACUACUUUAGUCGAGAUUACUAAUACAAUUCACUU ..((((.(((((((((((...(((((((.......))))))))))))))).(((((..((-(.....))).)))))..........))).)))).................. ( -26.60, z-score = -2.81, R) >consensus AUACUUGGGCAAGGAUCACGAAGGUUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGUUGGUUAUAAAAUAUAUUUCAAGUUUAAGAAUGUUAUUUAAUCACUUAAAUC_ ....((((((............((((((.......)))))).((((((........))))))...................))))))......................... (-15.32 = -14.80 + -0.52)
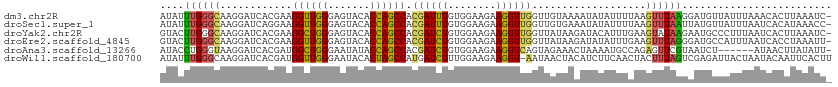
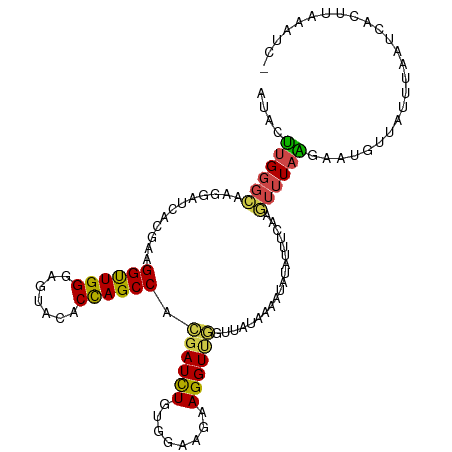
| Location | 12,681,439 – 12,681,547 |
|---|---|
| Length | 108 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.51 |
| Shannon entropy | 0.58751 |
| G+C content | 0.45929 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -17.63 |
| Energy contribution | -16.50 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.59 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904405 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 12681439 108 - 21146708 --------UUAAUUUAAAGGUUGGCAUUCCCGGAGAAAAAUAUUUGGGCAAGGAUCACGAAGGUUGGGAGUACACCAGCCACGAUUUGUGGAAGAAGGUUGGUUGUAAAAUAUAUU---- --------....((((....(..((...((((((........))))))......(((((((((((((.......))))))....)))))))......))..)...)))).......---- ( -23.20, z-score = -0.59, R) >droSim1.chr2R 11415478 108 - 19596830 --------UUAAUUUGAAGGUUGGCAUUCCCGGUGAAAAAUAUUUGGGCAAGGAUCACGAAGGUUGGGAGUACACCAGCCACGAUUUGUGGAAGAAGGUUGGUCGUAAAAUAUAUU---- --------...((((...((..(((...(((((..........)))))......(((((((((((((.......))))))....)))))))......)))..))...)))).....---- ( -23.50, z-score = -0.06, R) >droSec1.super_1 10186233 108 - 14215200 --------UUAAUUUAAAGGUUGGCAUUCCCGGUGAAAAAUAUUUGGGCAAGGAUCAGGAAGGUUGGGAGUACACCAGCCACGAUUUGUGGAAGAAGGUUGGUUGUGAAAUAUAUU---- --------..........((((((.(((((((.(.(((....))).).)...((((.....))))))))))...)))))).((((((........))))))...............---- ( -21.80, z-score = 0.25, R) >droYak2.chr2R 4843208 108 + 21139217 --------UUAAUUUCUAGGUUGGCAUUCCCGGUGAAAAGUACUUGGGCAAGGAUCACGAAGGCUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGUUGGUUAUAAGAUACAUU---- --------......(((.((((((.(((((((((.....((.(((....)))....))....)))))))))...)))))).((((((........))))))......)))......---- ( -28.70, z-score = -0.76, R) >droEre2.scaffold_4845 9418748 107 + 22589142 --------UUAA-UUCUAGGUUGGCAUUCCCGGUGAAAAGUACUUGGGCAAGGAUCACGAAGGUUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGUUGGUUAUAAGAUAUAUU---- --------....-.(((.((((((.(((((((..........(((....)))....((....)))))))))...)))))).((((((........))))))......)))......---- ( -25.60, z-score = -0.16, R) >droAna3.scaffold_13266 5970198 104 + 19884421 ------------UUUUUAGGUUGGUAUUCCUGGAGAGAAAUACCUGGGUAAGGAUCACGAUGGCUGGGAAUAUACCAGCCACGAUCUGUGGAAGAAGGUCAGUAGAAACUAAAAUG---- ------------.((((((...((((((.((....)).))))))....((..((((.((.(((((((.......))))))))).(((.....))).))))..))....))))))..---- ( -30.40, z-score = -2.17, R) >dp4.chr3 11182519 102 - 19779522 -------UUCCCUCUUCAGGUUGGCAUUCCCGGCGAGAAGUUUCUGGGCAAGGACCACGAUGGUUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGUCAGUAGCAGC----------- -------..((.(((((.((((((.(((((((.(.(((....))).).)...((((.....))))))))))...))))))((.....)).))))).))...........----------- ( -32.30, z-score = -0.04, R) >droPer1.super_4 6505076 102 - 7162766 -------UUCCCUCUUCAGGUUGGCAUUCCCGGCGAGAAAUUUCUGGGCAAGGACCACGAUGGUUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGUCAGUAGCAGC----------- -------..((.(((((.((((....(((((((.((....)).))))).)).)))).((.(((((((.......))))))))).......))))).))...........----------- ( -33.00, z-score = -0.47, R) >droWil1.scaffold_180700 3751660 117 + 6630534 --UUAAUUCAUUCCUUUAGGUUGGCAUUCCCGGUGAGAAAUAUUUGGGCAAGGAUCACGAUGGUUGGGAAUACACUAGCCAUGAUCUUUGGAAGAAGGU-AAUAACUACAUCUUCAACUA --.....(((..((....)).)))....(((((..(....)..)))))(((((((((...(((((((.......))))))))))))))))(((((..((-(.....))).)))))..... ( -29.90, z-score = -1.18, R) >droVir3.scaffold_12875 1540598 87 - 20611582 ----------UGUUUGUAGGUUGGCAUUCCUGGUGAAAAAUAUCUGGGCAAGGAUCAUGAGGGUUGGGAGUAUACCAGUCACGAUCUGUGGAAGAAG----------------------- ----------..((..(((((((....((((.((((......(((......))))))).))))((((.......))))...)))))))..)).....----------------------- ( -21.00, z-score = -0.45, R) >droMoj3.scaffold_6496 4648935 88 - 26866924 ---------GUUUCUGUAGGUUGGCAUUCCUGGCGAGAAAUACUUGGGCAAGGAUCAUGAGGGCUGGGAGUACACCAGUCACGAUCUGUGGAAGAAG----------------------- ---------.((((..(((((((.(((((((.(((((.....)))..)).))))..)))..((((((.......)))))).)))))))..))))...----------------------- ( -29.50, z-score = -1.91, R) >droGri2.scaffold_15245 8864149 88 + 18325388 ---------AUUUCUUCAGGUUGGCAUUCCCGGCGAAAAGUUUUUGGGCAAGGAUCACGAGGGUUGGGAAUAUACUAGCCACGAUUUGUGGAAGAAG----------------------- ---------..((((((.((((((.(((((((((.....((((((....)))))).......)))))))))...))))))((.....)).)))))).----------------------- ( -23.80, z-score = -1.00, R) >anoGam1.chr3R 874730 97 + 53272125 GUACCCAGAAUGGAUAAAAGUUGGCGUUCCGGGUGAAAAAUAUCUCGGUAAAGAUCACGAUGGCUGGGAAUACACCAGCCACGAUCUGUGGAAGAAG----------------------- .(((((.(((((............))))).))))).......((((.(...(((((....(((((((.......))))))).))))).).).)))..----------------------- ( -29.20, z-score = -2.16, R) >apiMel3.Group7 9169749 83 + 9974240 --------------UCAAGAUCGGCAUUCCCGGAGAACAUUAUCACGGUACCGAUCAUCAUGGAUGGCAGUAUACCAGUCAUGAUCUGUGGAAGAAG----------------------- --------------....((((((.....(((((........)).)))..)))))).(((((..(((.......)))..)))))(((.....)))..----------------------- ( -22.60, z-score = -1.07, R) >consensus ________UUAAUUUUUAGGUUGGCAUUCCCGGUGAAAAAUAUUUGGGCAAGGAUCACGAUGGUUGGGAGUACACCAGCCACGAUCUGUGGAAGAAGGU__GU_G_AA____________ .................((((((.....(((((..........))))).............((((((.......)))))).))))))................................. (-17.63 = -16.50 + -1.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:28:09 2011